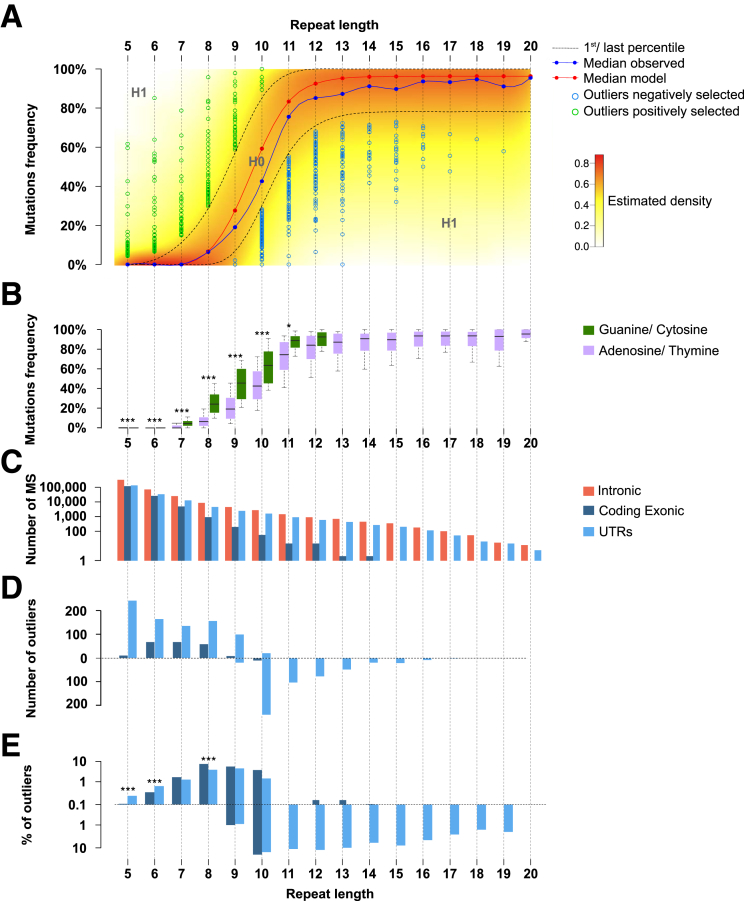
Figure 4.
Positive and negative selection in repetitive sequences in MSI CRC. (A) Distribution model of mutation frequencies across samples in microsatellites in UTRs or coding exonic regions according to repeat length for A/T nucleotides. The color gradient indicates the density of the β-binomial logistic regression model. The blue curve represents the median of observed mutation frequencies, and the red curve represents the median obtained from the model. This figure shows the statistical model and outliers for adenosine/thymine. (B) Box plot representation of mutation frequency variations according to the nucleotide composition of the microsatellite and to the repeat length. The significant independence of the chi-squared distribution is annotated by asterisks as follows: *P < .05, and ***P < .001. (C) Distribution of microsatellite mutations (log10 scale) in the 3 gene regions (UTRs, coding exonic, and intronic) according to repeat length. (D) Distribution of outlier mutation in microsatellites contained in UTRs and in coding exons. Positively and negatively selected microsatellite mutations are represented above and below the dotted line, respectively. (E) Distribution of the percentage of outlier mutations (log10 scale) according to repeat length. The significant independence of the chi-squared distribution is annotated by asterisks, as follows: *P < .05, **P < .01, and ***P < .001.