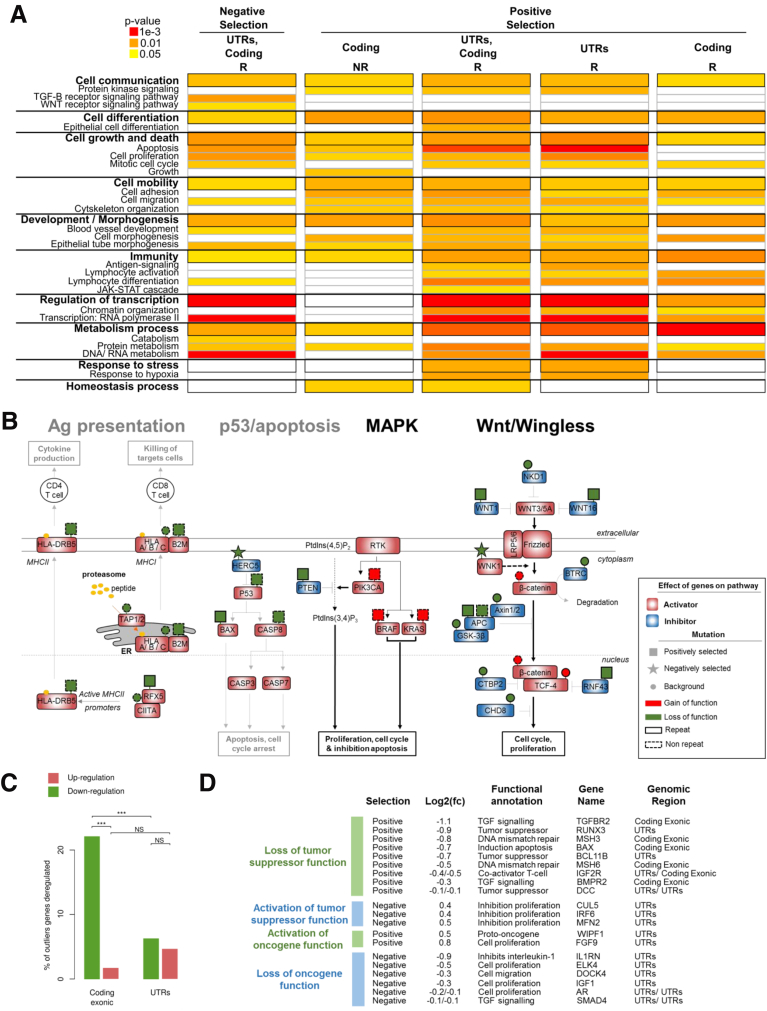
Figure 7.
Participation of positively and negatively selected events in cancer-related pathways. (A) Significant gene ontology terms enriched in outlier mutations compared with their abundance in all genes in the genome. Details of all gene ontology terms are available in Supplementary Table 4. (B) Interactions between mutations in R (continuous outlines) and NR sequences (dotted outlines) result in a coordinated effect on 4 signaling pathways. The effects of mutations that are unselected (circles), positively selected (rectangle), or negatively selected (stars) are indicated above the genes by a color code (red for gain of function and green for loss of function). Prediction of the global effect of mutations is represented by bold font for activation of the pathway and light grey font for inhibition. Genes known to be positive regulators (activators) of signaling pathways are indicated by a red rectangle and negative regulators (inhibitors) are indicated by a blue rectangle. (C) Percentage of outlier mutations in R sequences located in coding exons and in UTRs leading to significant dysregulation of mRNA expression level (comparison of mutated and wild-type tumors in each case). Upper: Proportion of microsatellite mutations leading to up-regulation of gene expression (in brown); lower: Down-regulation (in green). The significant independence of the chi-squared distribution is annotated by asterisks, as follows: ***P < .001. (D) List of MS candidate outliers that may contribute to the MSI tumor phenotype because of their biological functions, the effect of mutations on the level of gene expression, and the selective pressure at which the MS is constrained (positive or negative selection). Log2(fold change (Log2(fc))) indicates the log base 2 of mRNA expression between the MS mutant allele vs MS wild type. For each gene, the selective pressure, biological function, gene name, and the gene region of the MS are indicated.