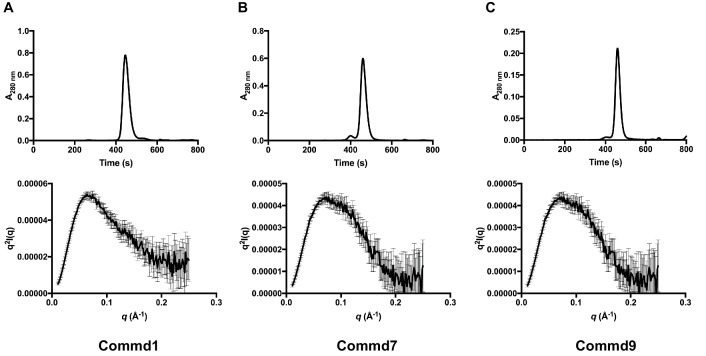
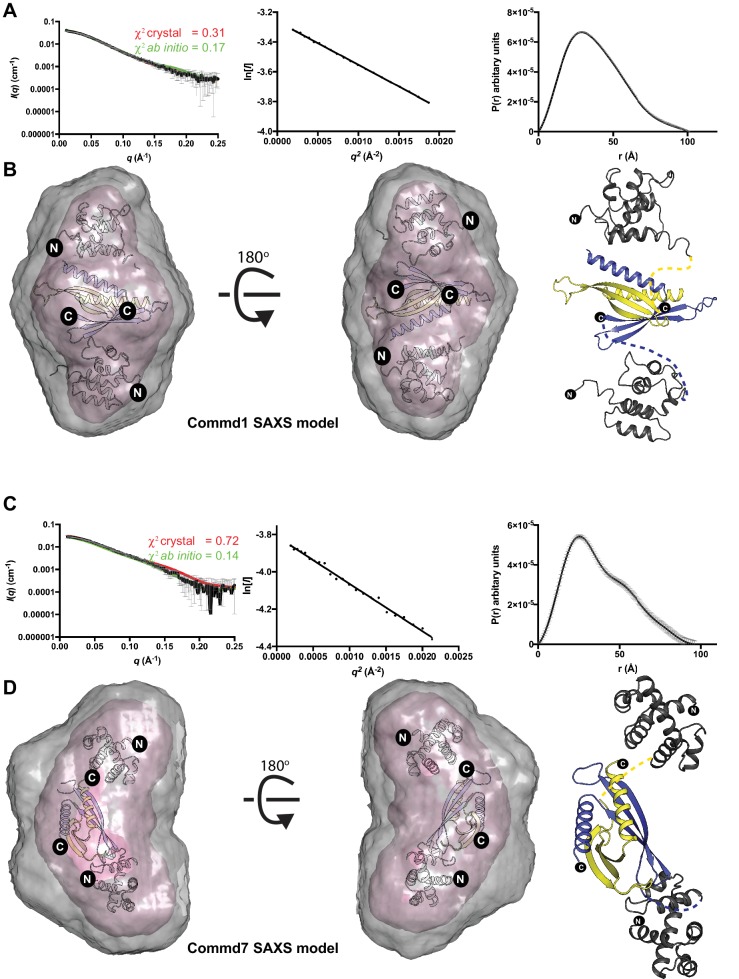
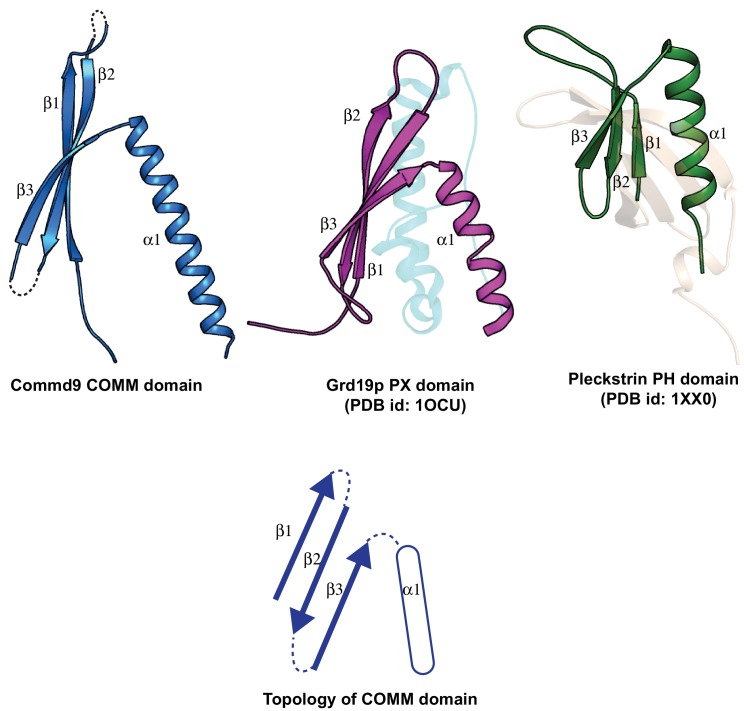
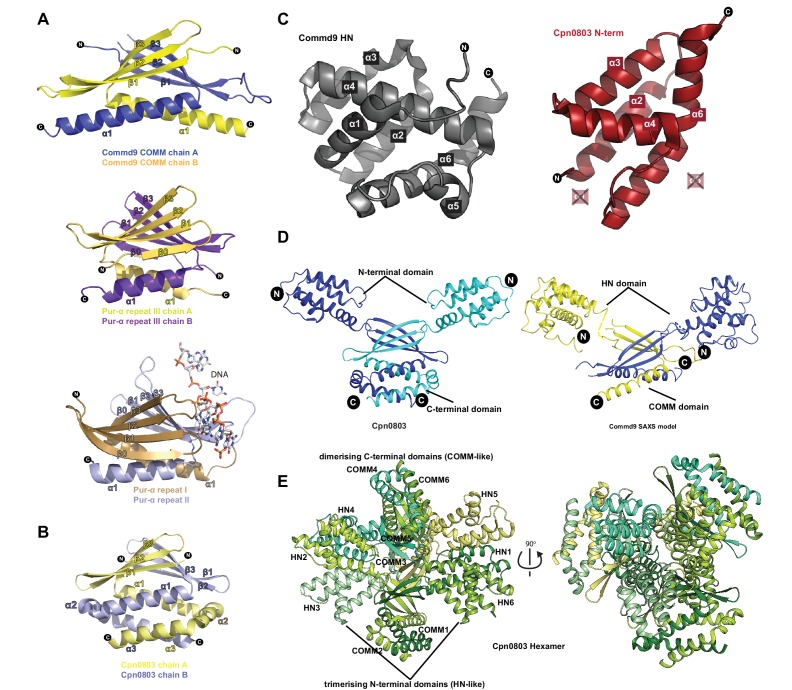
Figure 4. The solution structure of Commd9 determined by SAXS.
(A) (Left) Experimental scattering profile Commd9 in black overlaid with the theoretical scattering curve calculated from the Commd9 rigid body model (red) and the ab initio model determined by GASBOR (green) using CRYSOL. (Middle) The Guinier plot for the experimental data at the low-angle region (qmax × Rg < 1.3). (Right) P(r) functions derived from the SAXS data. (B) (Left) Averaged (grey) and filtered (coral) molecular envelopes from GASBOR. The ab initio model of Commd9 was docked with the rigid body model of Commd9 using SUPCOMB. (Right) Model of Commd9 protein from the experimental SAXS shown in ribbon diagram with HN domains in grey and the COMM domain dimer in blue and yellow.