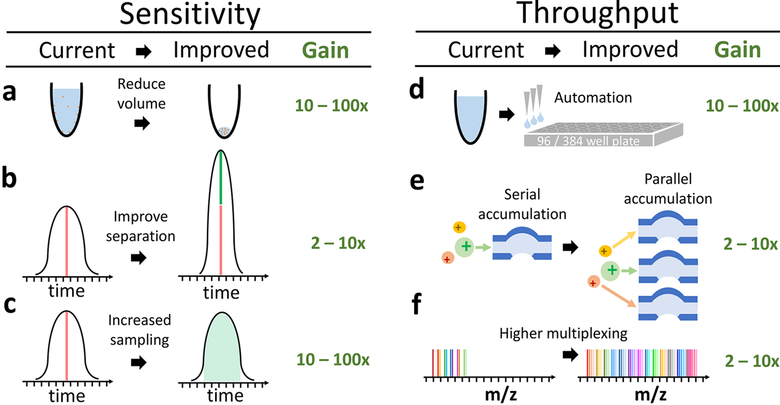
Figure 3.
Transformative opportunities for improving the quantification of single-cell proteomes. (a) Most bulk samples prepared for MS have volume of 10−100 μL.11,12,17 Reducing the volume for sample preparation to 1 to 2 nL13 can significantly reduce protein losses from surface adsorption. (b) The sharper the separation peaks, the larger the fraction of the ions can be analyzed for a fixed sampling (injection) time. Sharper peaks can be achieved by reducing the bore of LC columns, using monolithic columns, PLOT columns,27 or capillary electrophoresis.25 (c) Typically elution peaks have a full width at the base of ∼60 s and about 10−15 s at midheight, whereas ions for MS2 are sampled for mere milliseconds. These settings are typical for bulk proteomics and result in sampling <1% of the ions delivered to the instruments. Thus increasing the sampling time 100× can substantially increase the ions analyzed by MS, the sensitivity, and the accuracy of quantification. While, the panel displays sampling during the apex of the peak, this cannot always be achieved for all ions. (d) Automated liquid handling and 96/384-well plates can increase the consistency of sample preparation, decrease volumes to the nanoliter range, and increase throughput. (e) Parallel accumulation and serial injection of ions can afford increased ion sampling without reducing throughput. (f) A larger number of barcodes will increase the number cellular proteomes quantified per run without reducing proteome coverage or ion sampling.