FIGURE 1.
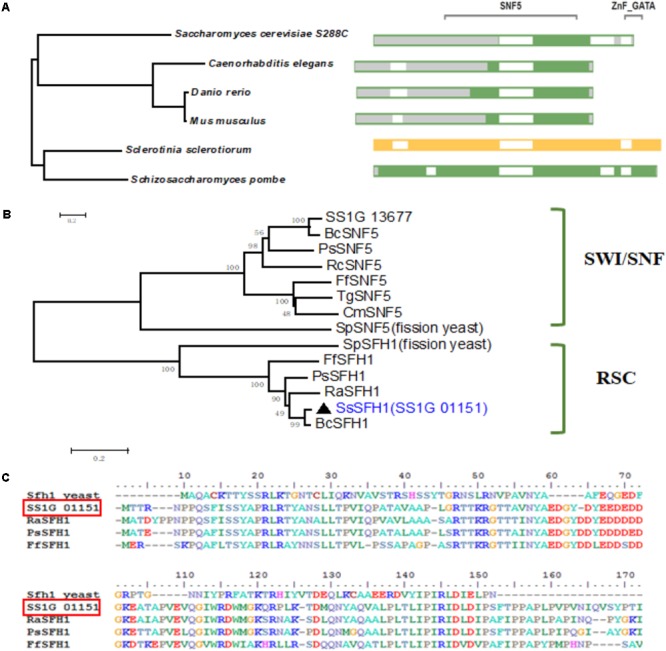
Sequence and phylogenetic analysis of SsSFH1. (A) SmartBLAST with NCBI: The query sequence SsSFH1 was highlighted in yellow; the amino acids, which are not aligned to the query sequence are shaded in gray. The gap is in white; landmark matches are shaded in green. Sequences are from S. sclerotiorum, Danio rerio, Mus musculus, Schizosaccharomyces pombe, Saccharomyces cerevisiae S288c and Caenorhabditis elegans. (B) Phylogenetic analysis of the conserved domain from different species. The selected proteins are: S. sclerotiorum, SsSFH1 (XP_001596958.1) and SS1G_13677 (XP_001585438.1); Fusarium fujikuroi, FfSFH1 (KLP21684.1) and FfSNF5 (KLP22090); Botrytis cinerea, BC1G_12110 (XP_001549133.1) and BcSNF5 (XP_001546059.1); Phialocephala subalpina, PsSFH1 (CZR60918.1) and PsSNF5 (CZR66927.1); Rhynchosporium agropyri, RaSFH1 (CZT00122.1); Rhynchosporium commune, RcSNF5 (CZT07508.1); Cordyceps militaris, CmSNF5 (ATY60809.1); Trichoderma guizhouense, TgSNF5 (OPB42243.1); and Schizosaccharomyces pombe, SpSFH1 (NP_588001.1) and SpSNF5 (NP_592979.1). The proteins are grouped into fungal SWI/SNF and RSC. Phylogenies were speculated using MEGA (version 7.05) program with the Neighbor-Joining method (1000 bootstrap replicates) to create an unrooted phylogenetic tree. (C) Sequence alignment of SNF5 domain from representative SFH1 proteins: S. sclerotiorum, SsSFH1 (XP_001596958.1), Fusarium fujikuroi, FfSFH1 (KLP21684.1), Phialocephala subalpina, PsSFH1 (CZR60918.1), Rhynchosporium agropyri, RaSFH1 (CZT00122.1), and Schizosaccharomyces pombe, Sfh1 (NP_588001.1). The sequence alignment was performed with BioEdit program.
