FIGURE 6.
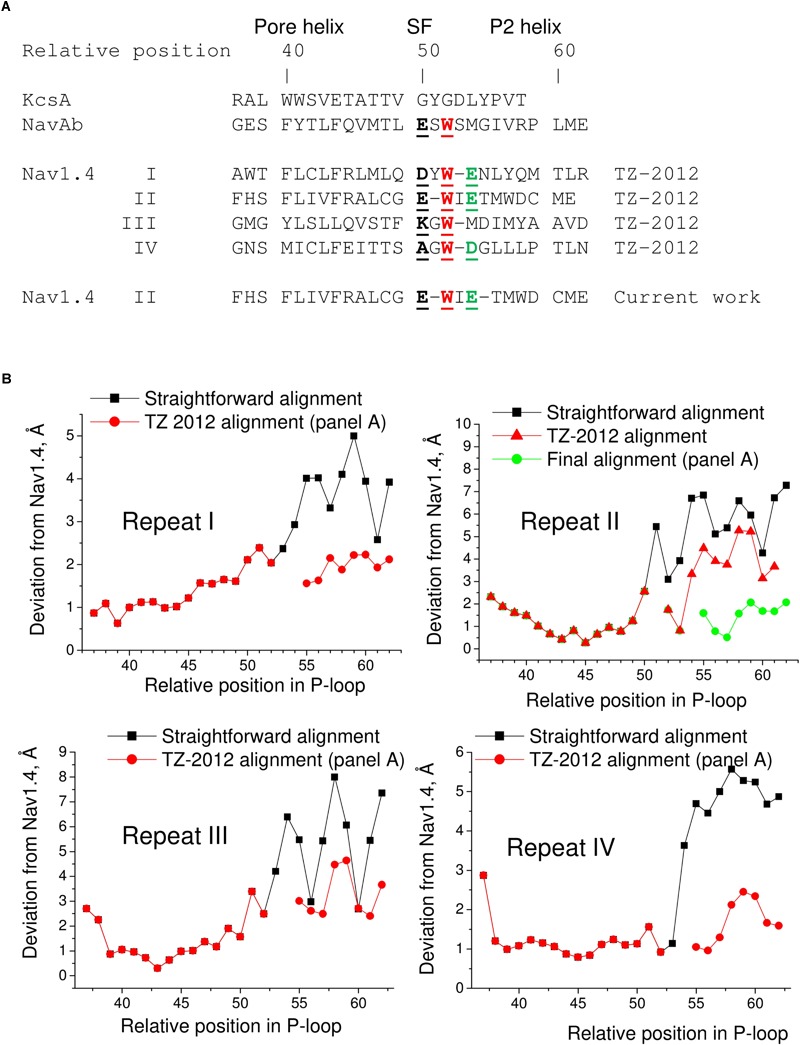
Adjusted sequence alignment decreases deviations of matching atoms in 3D aligned structure of NavAb. (A) Adjusted sequence alignment (TZ-2012) of P-loops in KcsA, NavAb, and Nav1.4 (Tikhonov and Zhorov, 2012). Highlighted are residues in the DEKA locus, conserved tryptophans that stabilize folding of the P-loops, and the outer carboxylates. In the adjusted, but not the straightforward (Figure 5A) sequence alignment the conserved tryptophans are in the matching positions. The second deletion in repeat II of Nav1.4 is introduced in the current work to minimize deviations between matching alpha carbons in the P2 helices of NavAb and Nav1.4. (B) Deviation of alpha carbons in repeats I-IV of Nav1.4 from the NavAb atoms, which are considered as matching according different sequence alignments. Introducing deletions decreases deviations and smoothes the deviation curves.
