FIGURE 7.
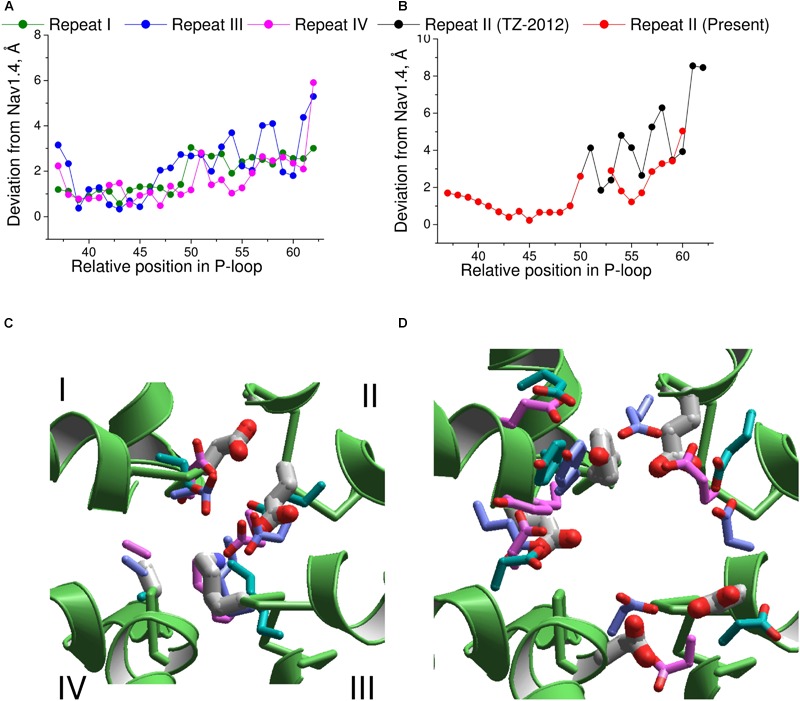
Comparison of the Nav1.4 cryo-EM structure with homology models. (A) P-loop alpha carbons in repeats I, III, and IV of the NavAb-based model (Tikhonov and Zhorov, 2012) are close to the matching atoms in the Nav1.4 structure. (B) In repeat II, big irregular deviations are seen because the Nav1.4 sequence aligned with NavAb lacks deletion at position 54 (Figure 6A). Introducing this deletion reduced deviations and the curve becomes similar to those in other repeats. (C) Side chains of the DEKA ring in 3D aligned structures. Only the Nav1.4 backbone is shown. The Nav1.4 residues are shown as thick sticks. Residues in the KcsA-based models (Lipkind and Fozzard, 2000; Tikhonov and Zhorov, 2005a) and NavAb-based model (Tikhonov and Zhorov, 2012) are shown, respectively, with cyan, magenta, and violet thin sticks. (D) Side chains of the outer carboxylates and a TTX-interacting tyrosine in position 51 of repeat I. Residues are rendered as in (C).
