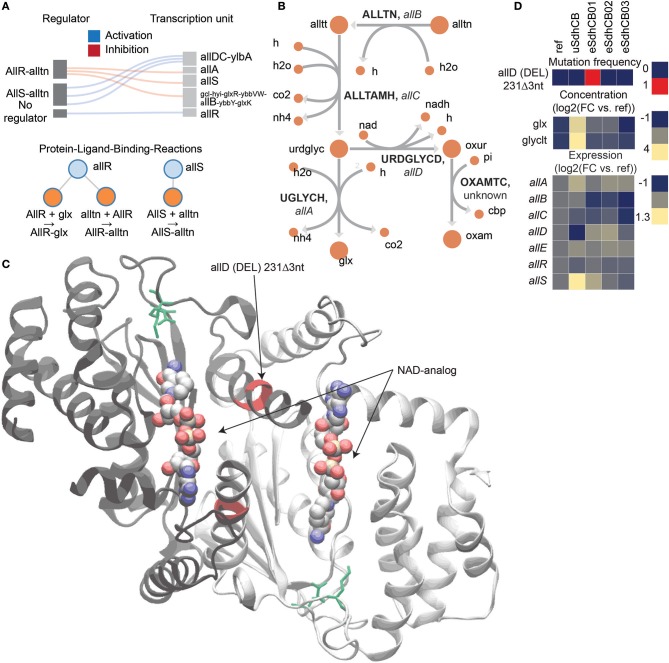
Figure 5.
A mutation in the active site of ureidoglycolate dehydrogenase (URDGLYCD) in eSdhCB01 that potentially provided an auxiliary means to metabolize excess glyoxylate (glx) and/or balance nitrogen levels. (A) Regulatory diagram and protein-ligand-binding reactions. The allDCE operon is positively regulated by AllS, whose gene expression is negatively regulated by AllR (Rintoul et al., 2002). allA and allB are also negatively regulated by AllR (Rintoul et al., 2002). Glyoxylate enzymatically inhibits AllR repression of allS, allA, and allB (Rintoul et al., 2002). (B) Network diagram of allantoin assimilation. (C) Crystal structure of AllD. Mutations are highlighted in red. Active site residues are highlighted in green. An NAD-analog where NAD would bind is shown. Note that the deletion (DEL) mutation in eSdhCB01 would affect active site residues, and most likely binding of NAD and other substrates. (D) Mutation frequency, metabolite levels, and gene expression of components involved in allantoin assimilation. Note that gene expression profiles are consistent with high glyoxylate levels in uSdhCB.