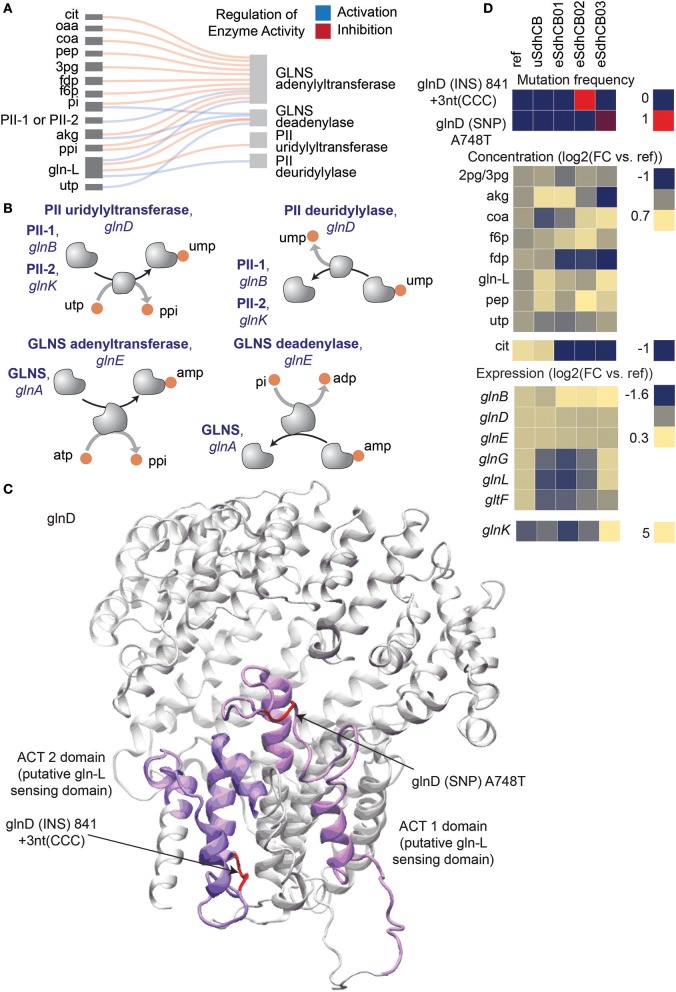
Figure 6.
Mutations that potentially affect regulation of nitrogen assimilation in eSdhCB strains. Mutations in glnD, which encodes the primary nitrogen status sensor PII uridylyl-/deuridylyl-transferase, fixed in eSdhCB02 and overtook a majority of the population in eSdhCB03. (A) Regulatory diagram of allosteric regulation of PII uridylyl-/deuridylyl-transferase and GLNS adenylyltransferase/GLNS deadenylase. (B) Protein-protein interaction diagram of PII uridylyl-/deuridylyl-transferase and GLNS adenylyltransferase/GLNS deadenylase. Increased levels of gln-L increase the deuridylylation activity of glnD and decrease the uridylyltransferase activity of glnD (Jiang et al., 2012). PII-ump stimulates deadenylation of GLNS via glnE while PII stimulates adenylation of GLNS (Rhee et al., 2006; van Heeswijk et al., 2013), the removal of amp enhances GLNS activity (Rhee et al., 2006; van Heeswijk et al., 2013). (C) Crystal structure of PII uridylyl-/deuridylyl-transferase [Reference]. Mutations are highlighted in red. The ACT 1 and 2 domains are highlighted in dark and light magenta, respectively. The ACT 1 and 2 domains are believed to play a role in sensing the intracellular levels of gln-L (Zhang et al., 2010; Jiang et al., 2012). Note that the mutations were located in the ACT 1 and 2 domains. (D) Mutation frequency, metabolite levels, and gene expression levels of components involved in E. coli nitrogen assimilation.