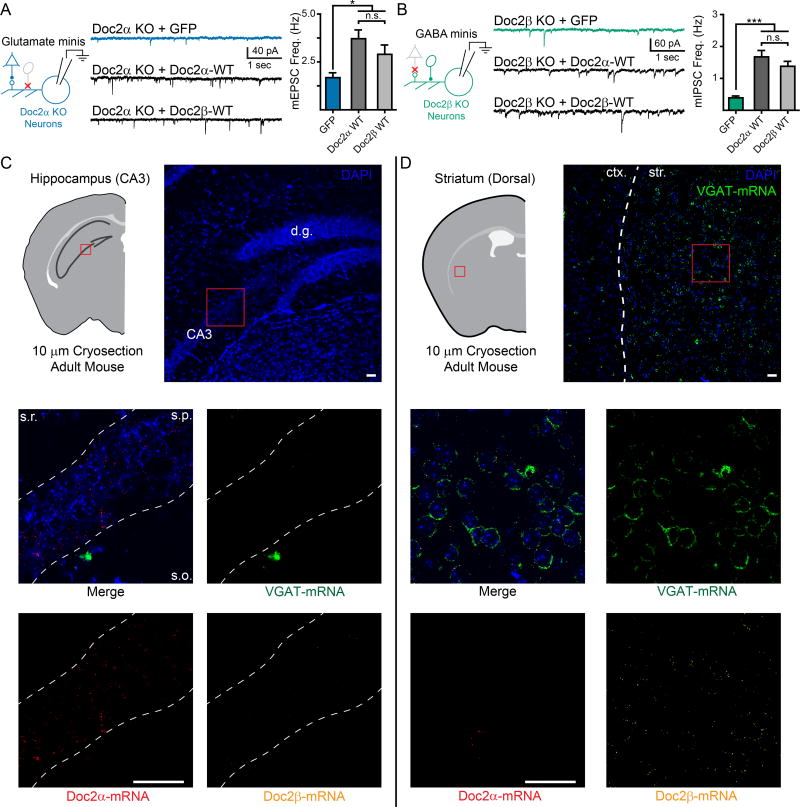
Figure 5.
Doc2α and β are functionally redundant but differentially expressed in excitatory and inhibitory neurons. A. Representative traces (center) and quantification (right) demonstrating that neuronal, non-subtype specific expression of Doc2β WT rescues mEPSC frequency in Doc2α single KO neurons (left). B. Representative traces (center) and quantification (right) demonstrating that neuronal, non-subtype specific expression of Doc2α WT rescues mIPSC frequency in Doc2β single KO neurons (left). Note, the experiments presented in panels A and B were performed in parallel to those in Fig. 4B and D, respectively. Thus, the data for GFP controls, Doc2α WT in panel (A), and Doc2β WT in panel (B), are reproduced here from those previous figures to facilitate comparisons. C and D. RNAScope images examining the cellular distribution of Doc2 isoforms in glutamatergic CA3 pyramidal neurons in the hippocampus (C) and GABAergic neurons in the striatum (D) visualized in coronal brain slices (10 µM thick) obtained from adult mice. Upper. Cartoon representation and low-magnification images demonstrating gross morphology of the brain slices imaged. Abbreviations: d.g. = dentate gyrus, ctx. = cortex, and str. = striatum. Red boxes approximate the regions examined in the lower panels, but do not directly correlate. Middle and lower. High-magnification images showing the distribution of Doc2α-mRNA (red), Doc2β-mRNA (yellow), and VGAT-mRNA (green) among neurons (indicated by DAPI, blue). Abbreviations: s.r. = stratum radiatum, s.p. = stratum pyramidale, and s.o. = stratum oriens. Scale bars (white) represent 50 µm and apply to all lower panels. Images were adjusted for brightness/contrast; corresponding channels underwent the exact same adjustments. RNAScope experiments were performed three times, on separate animals, and similar results were observed each time. White dashed lines were overlaid on the images to help highlight morphology. *** indicates p < 0.001, * indicates p < 0.05, and n.s. indicates p > 0.05. Bar graphs represent mean ± SEM.