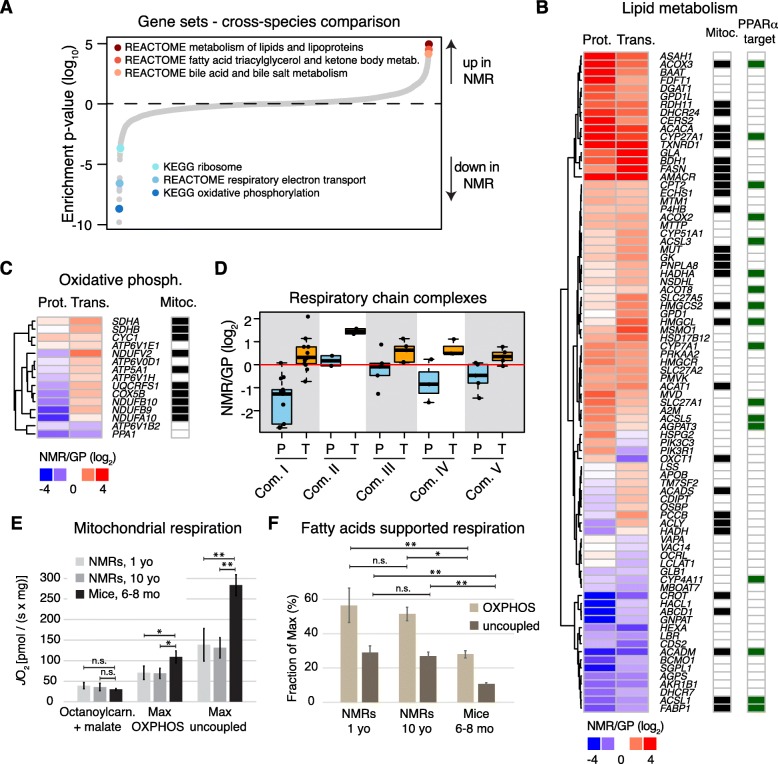
Fig. 2.
Distinctive features of NMR liver metabolism. a Gene set enrichment analysis was performed on proteomic data. Gene sets are plotted according to the log10 value of the calculated enrichment score. Positive and negative values are used for gene sets showing higher and lower abundance in NMR, respectively. Because of the redundancy among the significantly affected gene sets (q < 0.05), we highlighted representative categories of gene sets. The complete list of enriched gene sets is available in Additional file 4: Table S3. b Protein (Prot.) and transcript (Trans.) fold changes for genes involved in lipid metabolism (“REACTOME metabolism of lipids and lipoproteins”, combined RNA-seq and proteome q < 0.001), and c oxidative phosphorylation that are significantly affected in NMR vs. GP (selection criteria as in b). Genes encoding for proteins localized to mitochondria and targets of PPARα are indicated in black and green in the annotation heat maps, respectively. d Fold change comparison for genes of the different complexes of the respiratory chain. Light blue and orange boxes indicate protein and transcript fold changes, respectively. e Mitochondrial oxygen flux supported by octanoylcarnitine and malate, compared to maximum coupled (OXPHOS) and maximum uncoupled respiration in liver of NMR (1 and 10 years old) and 6–8-month male mice. f Ratio of fatty acid supported respiration to maximum OXPHOS and uncoupled respiration in the liver of NMR and mouse. In e and f, reported values are averages obtained from n = 4 animals per experimental group ± standard deviation. *p < 0.05; **p < 0.001; n.s = not significant. Related to Additional file 4: Table S3