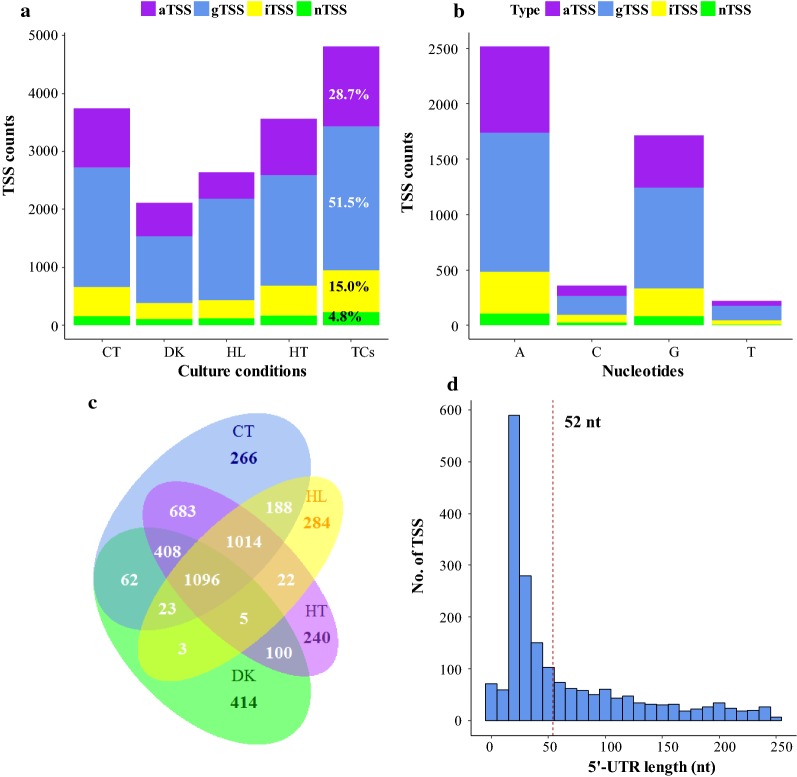
Fig. 1.
Statistical analysis of the TSSs identified in the transcriptomes of S. elongatus UTEX 2973. TSSs with ≥ 300 raw reads were considered to be active and were included in the following statistical analysis. TSSs were classified into four different types according to previous studies [21, 22]: gTSS (blue), aTSS (purple), iTSS (yellow), and nTSS (green). a Numbers of each type of TSS identified from the different growth conditions. CT, DK, HL and HT represent control, dark, high-light and high-temperature conditions, respectively. TCs represent the total of all TSSs identified across the four conditions. b Nucleotide usages for each TSS type. c Distribution of TSSs active under different respective growth conditions. d Histogram showing the distribution of 5′-UTR lengths of gTSSs. The median 5′-UTR length is indicated as a dashed line