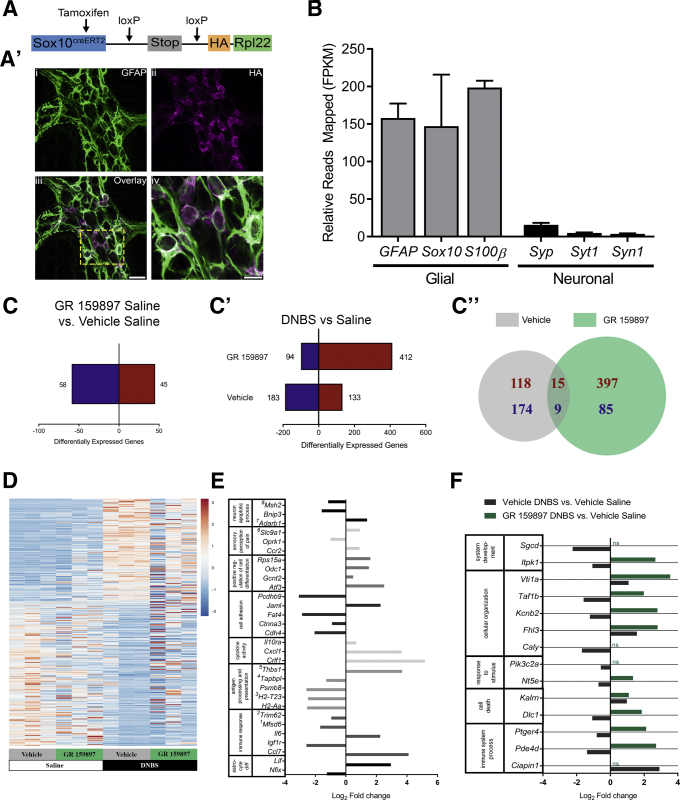
Figure 10.
Glial transcriptome alteration driven by acute inflammation. (A) Schematic of transgene containing the HA-tagged ribosomal protein L22 (Rpl22) driven by the tamoxifen inducible glial promoter (SOX10creERT2). (A’) Confocal image of dual-label immunohistochemistry (single optical plane, 1 μm) for the HA-tagged Rpl22 (magenta) and enteric glia (GFAP, green) in the myenteric plexus of the Sox10::CreERT2+/-/Rpl22tm1.1Psam/J mouse distal colon. (A'iii) The area marked is enlarged in panel A'iv. (A'iii) Scale bar: 20 μm (applies to A'i–iii). (A'iv) Scale bar: 10 μm. (B) Mean fragments per kilobase of transcript per million mapped reads (FPKM) values of sentinel glial and neuronal genes from vehicle saline samples showing increased relative expression of glial to neuronal transcripts (n = 3 mice; data shown represent biological replicates). (C) Total numbers of differentially expressed enteric glial genes in saline mice treated with the NK2R antagonist GR 159897 or vehicle (GR 159897 saline compared with vehicle saline; n = 3 mice; P < .005; data shown represent biological replicates). (C’) Total numbers and (C”) Venn diagram comparison of differentially expressed enteric glial genes 48 hours after induction of DNBS colitis in mice treated with the NK2R antagonist GR 159897 or vehicle compared with vehicle saline mice. (C–C”) Up-regulated genes are shown in red and down-regulated genes are shown in blue. (D) Heatmaps for all 316 differentially expressed genes in the vehicle DNBS vs vehicle saline comparison in order of fold-change (positive to negative) for all groups (n = 3 mice; P < .005; data shown represent biological replicates). Color scale is based on gene-based Z-scores of log2 FPKM values. (E) Selected differentially expressed genes classified by inflammation-related gene ontology (GO) terms from the distal colon of vehicle DNBS mice compared with vehicle saline mice (n = 3 mice; P < .005; data shown represent biological replicates). Superscripts denote actual categorization in similar, more specific GO terms, which were grouped together under a broader category of GO terms, as follows: 1adaptive immune response; 2innate immune response in mucosa; 3antigen processing and presentation of peptide antigen via major histocompatibility complex class I; 4negative regulation of antigen processing and presentation of peptide antigen via major histocompatibility complex class I; and 5negative regulation of peptide or polysaccharide antigen via major histocompatibility complex class II. (F) Selected differentially expressed genes classified by biological process GO categories, which are significantly differentially regulated between GR 159897 DNBS and vehicle DNBS mice (n = 3 mice; P < .005; data shown represent biological replicates). Relative changes shown are compared with vehicle saline.