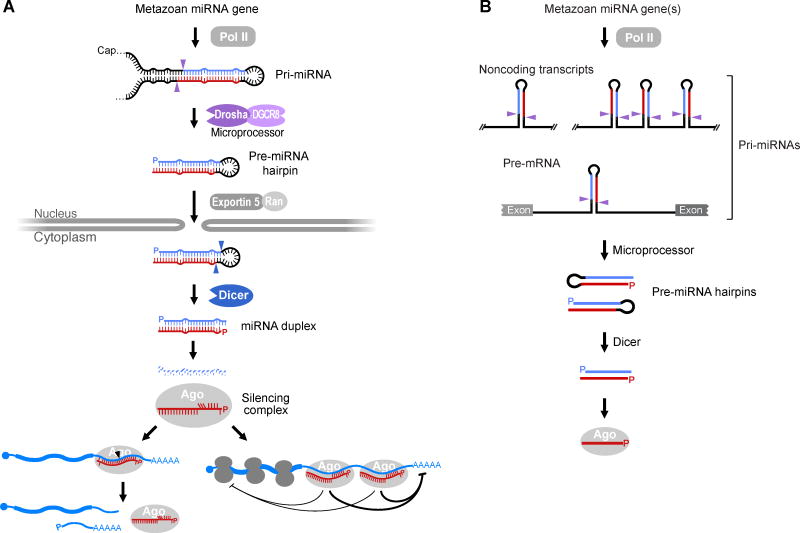
Figure 1. The Biogenesis and Function of Canonical miRNAs of Animals.
(A) Biogenesis and function of a typical miRNA. Once transcribed by Pol II, the pri-miRNA folds back on itself to form at least one distinctive hairpin structure (further described in Figure 2), which is cleaved by Microprocessor (purple arrowheads, cleavage sites) to release a pre-miRNA (P, 5′ phosphate). The pre-miRNA hairpin is exported to the cytoplasm through the action of Exportin 5 and RAN–GTP. In the cytoplasm, the pre-miRNA is cleaved by Dicer (blue arrowheads, cleavage sites) to produce a ~20-bp miRNA duplex with a 5′ phosphate (P) and a 2-nt 3′ overhang on each end. One strand of the miRNA duplex, the mature miRNA (red), is loaded into the guide-strand channel of an Argonaute protein (Ago) to form a silencing complex, whereas the other strand, the miRNA* (blue), is degraded. Within the free silencing complex, miRNA nucleotides 2–5 (upward red vertical lines) are poised to initially interact with target RNAs (blue; filled circle, cap; AAAAA, poly(A) tail). This pairing usually extends to nucleotide 7 or 8 of the miRNA, and occasionally is more extensive (Figures 5 and 6). If pairing is very extensive, the target can be sliced (left; black arrowhead, cleavage site), whereas if it is not, the target can undergo other types of repression (right; further described in Figure 4).
(B) Typical sources of canonical miRNAs. Most canonical miRNAs derive from the introns or exons of non-coding primary transcripts, some of which harbor hairpins for more than one miRNA. In addition, many canonical miRNAs derive from introns of pre-mRNAs.