Figure 2. The Menu of Structural and Primary-Sequence Features that Define Canonical miRNA Hairpins.
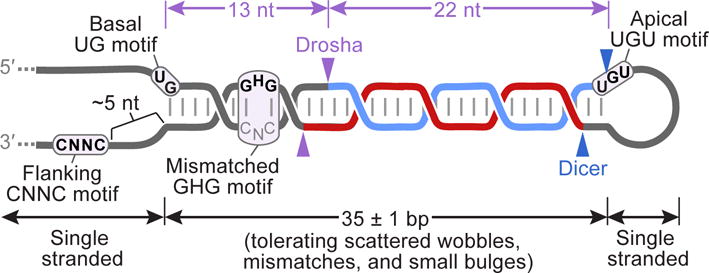
Structural features include a 35-bp stem with an unstructured loop and unstructured flanking regions. At every position within the stem except for one, pairing is preferred; this is typically Watson–Crick pairing, although at many positions one or both of the G–U wobble pairs is as favorable as some of the Watson–Crick pairs. Natural miRNA hairpins typically also have a few mismatches or small bulges scattered at various locations within the stem, and the mismatches are each counted as base pairs when describing the optimal length of 35 ± 1 bp. Additional features can enhance processing and help specify the sites of cleavage (purple arrowheads); these include a basal UG motif, an apical UGU motif, a flanking CNNC motif (in which N is any of the four nucleotides), and a mismatched GHG motif (in which H is A, C, or U), each located at the indicated positions relative to the Drosha cleavage sites (Auyeung et al., 2013; Fang and Bartel, 2015). Drosha recognizes the base of the hairpin, including the basal UG and probably the mismatched GHG motif, whereas the DGCR8 dimer recognizes the apical region, including the apical UGU (Nguyen et al., 2015), and together Drosha and DGCR8 form a molecular caliper that measures the length of the stem. An auxiliary factor, such as SRp20 or p72, recognizes the flanking CNNC motif (Auyeung et al., 2013; Mori et al., 2014). The sites of Dicer cleavage are also shown (blue arrowheads).
