Figure 5. MicroRNA Target Sites.
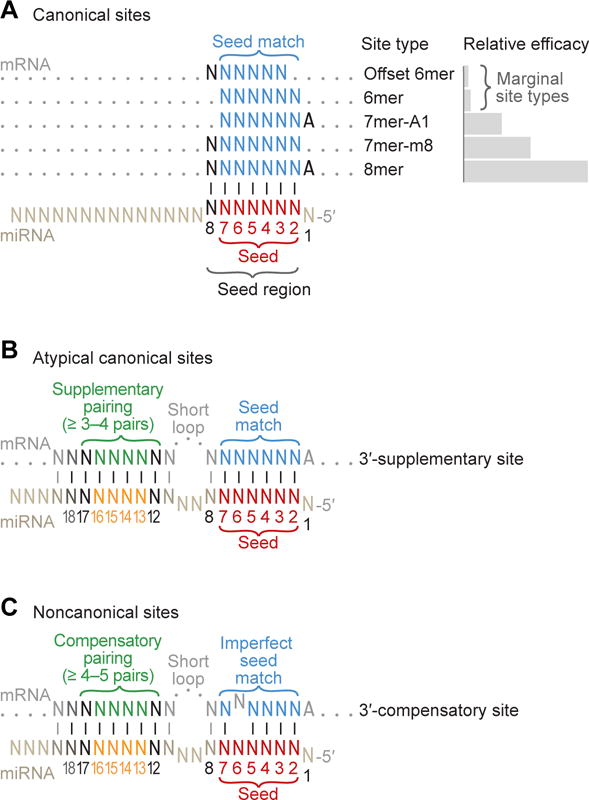
(A) Canonical sites of mammalian miRNAs. These canonical sites each have 6–7 contiguous Watson–Crick pairs (vertical lines) to the seed region of the miRNA (miRNA positions 2–8). Two of these sites also include an A at position 1. Relative site efficacy in mammalian cells is graphed to the right (log scale). The most effective canonical sites are 7–8 nt sites that include a perfect match to the miRNA seed (positions 2–7, red), whereas the 6 nt sites are the least effective. Not shown is the 6mer-A1 site (the same as the 7mer-A1 but lacking a pair at position 7), which is rarely conserved above background in mammalian mRNAs and has negligible efficacy in mammalian cells yet is sometimes included among the canonical sites because of its more robust conservation and efficacy in C. elegans (Jan et al., 2011).
(B) The 3′-supplementary site, an atypical type of canonical site. A small fraction of the canonical sites (< 5%) benefit from pairing to the 3′ region of the miRNA. Productive 3′-supplementary pairing typically centers on nucleotides 13–16.
(C) The 3′-compensatory site, a functional type of noncanonical site. The noncanonical sites do not have six contiguous Watson–Crick pairs to the seed region. Shown is a mismatch at position 6, but the imperfection (either a wobble, mismatch, or single-nucleotide bulge) can instead occur at another seed position. Compensating for the imperfect seed match is extensive pairing to the 3′ region of the miRNA, which typically centers on miRNA nucleotides 13–16.
