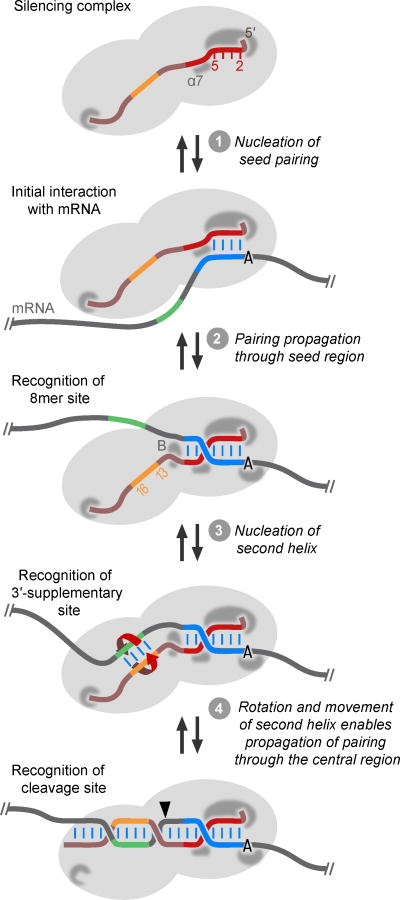
Figure 6. A Unified Model for miRNA Target Recognition and Pairing Propagation.
The miRNA is tightly bound within the silencing complex. A pocket within the Ago MID domain binds the miRNA 5′ terminus, and one within the PAZ domain binds its 3′ terminus. In addition, interactions with the seed region (red) pre-organize nucleotides 2–5 for initial pairing with the target (step 1). As pairing propagates through the seed region (step 2), α-helix 7 (α7), which initially imposes a kink in the seed and steric block to this propagation, shifts to a location at which it reinforces Watson–Crick paring at positions 6 and 7. Most canonical sites rely on this pairing to the seed region, often supplemented with an additional interaction between Ago and an A at target position 1, but with a steric and conformational block (B) preventing contiguous pairing from propagating beyond position 8. Some sites have a segment (green) that can pair to the miRNA 3′ region, optimally centering on miRNA nucleotides 13–16 (orange) to nucleate a second RNA helix (step 3). Because this second helix forms on the other side of the block, it can form without either steric hindrance or the conformational challenge of wrapping the miRNA around the mRNA. This second helix contributes the 3′-supplementary/compensatory pairing observed for some sites. For the very few sites that also have pairing through the center of the miRNA, pairing can then form between the second helix and the seed helix, propagating from either helix or from both simultaneously (step 4). To accommodate the torsional strain of forming this pairing between the helices, the second helix rotates around its helical axis, while the seed helix remains fixed. As the second helix begins to rotate and as pairing propagates from the second helix towards the miRNA 3′ terminus, the 3′ terminus is pulled from its binding pocket, relieving strain for further rotation of the second helix and full propagation of pairing (step 4). Propagation of pairing through the block and other RNA conformational changes that generate a long contiguous helix are coupled to protein conformational changes that help create the active site for target slicing (arrowhead). Each step prior to slicing is reversible, and thus this model suggests an analogous pathway for the stepwise release of the passenger strand during silencing-complex maturation, in which a miRNA duplex replaces the fully paired miRNA–target complex, and the pathway proceeds in reverse.