Abstract
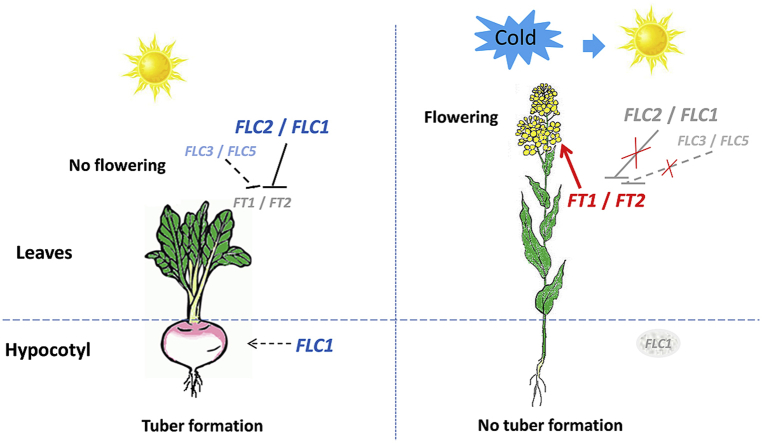
The turnip (Brassica rapa var. rapa) is a biennial crop that is planted in late summer/early fall and forms fleshy tubers for food in temperate regions. The harvested tubers then overwinter and are planted again the next spring for flowering and seeds. FLOWERING LOCUS C (FLC) is a MADS-box transcription factor that acts as a major repressor of floral transition by suppressing the flowering promoters FT and SOC1. Here we show that vernalization effectively represses tuber formation and promotes flowering in Tibetan turnip. We functionally characterized four FLC homologues (BrrFLC1, FLC2, FLC3, and FLC5), and found that BrrFLC2 and BrrFLC1 play a major role in repressing flowering in turnip and in transgenic Arabidopsis. In contrast, tuber formation was correlated with BrrFLC1 expression in the hypocotyl and was repressed under cold treatment following the quantitative downregulation of BrrFLC1. Grafting experiments of non-vernalized and vernalized turnips revealed that vernalization independently suppressed tuberization in the tuber or hypocotyl of the rootstock or scion, which occurred in parallel with the reduction in BrrFLC1 activity. Together, our results demonstrate that the Tibetan turnip is highly responsive to cold exposure, which is associated with the expression levels of BrrFLC genes.
Keywords: Tibetan turnip, Tuberization, Flowering, BrrFLC genes, Vernalization
Graphical abstract
1. Introduction
The turnip (Brassica rapa var. rapa), a member of B. rapa in the Brassicaceae family, is a biennial root crop that forms fleshy tubers, providing food for humans and livestock in temperate climates worldwide (Liang et al., 2006). Biennial turnips are planted in late summer/early fall and require vernalization during winter to flower the next spring. Controlling flowering time is especially important in turnip crops because early bolting can severely decrease the yield and quality.
Vernalization, the process by which exposure to cold prevents the premature flowering during warm autumn days, has a significant influence on crop yield (Kim et al., 2009). One of the key genes that regulates the vernalization requirement and response is FLC (FLOWERING LOCUS C), a MADS-box transcription factor that acts as a repressor of floral transition (Schmitz and Amasino, 2007, Sharma et al., 2017). FLC represses the flowering pathway integrators FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) (Michaels et al., 2005, Schmitz and Amasino, 2007). Prolonged cold causes epigenetic silencing of FLC, which releases FT and SOC1 from repression and promotes flowering after plants are moved to warm conditions (Bastow et al., 2004, De Lucia et al., 2008, Krichevsky et al., 2007). Association and quantitative trait loci (QTL) mapping studies have associated diversity in flowering time or vernalization response with polymorphisms in the FLC homologues of Brassica oleracea, B. rapa and Brassica napus (Hou et al., 2012, Irwin et al., 2016, Zhao et al., 2010). In monocots, FLC-like genes are found in cereals where they also respond to prolonged cold (Ruelens et al., 2013). Four FLC paralogues (FLC1, FLC2, FLC3 and FLC5) in B. rapa (Schranz et al., 2002) and in B. oleracea (Okazaki et al., 2007) have been cloned, and some of their molecular functions have been studied. For instance, one of two major FLC haplotypes in B. oleracea is repressed by cold exposure more slowly than the other (Irwin et al., 2016). In another study, a splicing site mutation in BrFLC1 has been shown to significantly contribute to diverse flowering times (Yuan et al., 2009). These findings indicate that in different genetic backgrounds, FLC genes play diverse roles on flowering time. Even though these FLC paralogues are apparently related to FLC, the extent of FLC conservation and the evolutionary versatility in B. rapa and other Brassica crops is still unclear.
In this study, we identified correlations among cold treatment length, tuberization inhibition, promotion of flowering, and decreased transcript levels in turnip BrrFLC homologues. The inhibition of vernalization on tuber formation was found to be concurrent with the silencing of BrrFLC1 gene expression in the hypocotyl. BrrFLC1 and BrrFLC2 in combination play a major role in repressing flowering and responding to vernalization.
2. Materials and methods
2.1. Plant materials and growth conditions
The Tibetan turnip (B. rapa var. rapa) is widely distributed in Tibet where it is traditionally used as a folk medicine and food to relieve hypoxia and alleviate fatigue (Chu et al., 2017). Tibetan turnip landrace KTRG-B17 was collected from Qüxü County, Lhasa, Tibetan Autonomous Region, China. Its seeds were sown in Petri dishes containing two pieces of filter paper in the dark at 22 °C for one day until germination. Germinated seedlings were planted in 0.1 L pots with a 3:1 ratio of soil and vermiculite. For the vernalization treatment, germinated seeds were incubated under a short day (SD) photoperiod at 5 °C for 10, 20, 30, 40, 50 or 60 days. Subsequently, the pots were moved to the greenhouse under long-day (LD) conditions with 16 h light/8 h darkness. Non-vernalized plants were transferred directly to the same conditions as vernalized plants.
2.2. Phenotyping analysis
Flowering time was measured as the number of days to flowering. The number of flowering plants was recorded on different days after vernalization. All measurements were based on 30 plants. Flowering times of Arabidopsis transgenic plants were recorded as the total number of leaves when the floral bolt was 1 cm high; 12 plants for each transgenic line were measured. The fleshy tuber diameter of the turnip was measured for the non-vernalization and vernalization treatments, respectively. Measurements were based on 30 plants.
2.3. RNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA was isolated using Trizol reagent (Invitrogen) from different tissues and growth periods. First-strand cDNA was synthesized from 1.5 μg of DNAse-treated RNA in a 20 μL reaction volume using M-MuLV Reverse Transcriptase (Invitrogen) with oligo (dT)18 primer. qRT-PCR was carried out using 2XSYBR Green I Master on a Roche LightCycle 480 real-time PCR machine in accordance with the manufacturer's instructions. At least three biological replicates and three technical replicates for each sample were used for qRT-PCR analysis. The BrrTUB2 gene and AtTUB2 genes were used as controls. Gene-specific primers are listed in Supplemental Table 1.
2.4. Construction of plant expression vectors and generation of transgenic plants
To generate BrrFLC1, FLC2, FLC3, and FLC5 constructs, cDNA was fused to GFP under the control of a cauliflower mosaic virus (CaMV) 35S promoter, the full-length cDNA coding region of the BrrFLC gene was amplified from the cDNA of Tibet turnip seedlings and then cloned downstream of the GFP fluorescence marker in the pEGAD vector between the EcoRI and BamHI sites. These constructs were directly transformed into Agrobacterium tumefaciens GV3101, and Col and Col-FRI-flc plants were transformed using Agrobacterium-mediated floral transformation, to generate the corresponding transgenic lines. Phenotyping was performed with T2 plants. Gene-specific primers are listed in Supplemental Table 1.
2.5. Protein immunoblotting
Total proteins were prepared by grinding seedlings on ice in an extraction buffer [50 mM Tris, 5% glycerol, 4% sodium dodecyl sulfate, 1% polyvinylpolypyrrolidone, 1 mM phenylmethylsulfonyl fluoride (pH 8.0)], followed by centrifugation at 14,000g at 4 °C for 15 min. The same amount of total proteins was loaded into a 10% SDS-PAGE gel and then transferred onto a PVDF blotting membrane, which was then probed with the appropriate primary anti-GFP (1:3000, Clontech) and horseradish peroxidase-conjugated goat anti-mouse secondary antibody (1:3000, Promega). Signals were detected using the ONE-HOUR IP-Western Kits (Cat. L00232, Genescript).
2.6. Graft method
The general grafting conditions and procedures were performed as previously reported (Marsch-Martínez et al., 2013). In brief, young vernalized and non-vernalized turnip seedlings were grown on ½ MS plates for 3–5 days. Under the dissecting microscope, the cotyledons were cut off from all seedlings using a scalpel. The scion seedlings were cut across the hypocotyl below the cotyledon stump to ensure that the rootstock seedlings were also aligned with their cotyledons flat. The stump of the rootstock and the cut stump of the scion were abutted so that the two phloem strands from the rootstock and scion matched. The successful graft lines were moved to the soil and grown under long-day conditions.
3. Results
3.1. Vernalization responses of Tibetan turnips
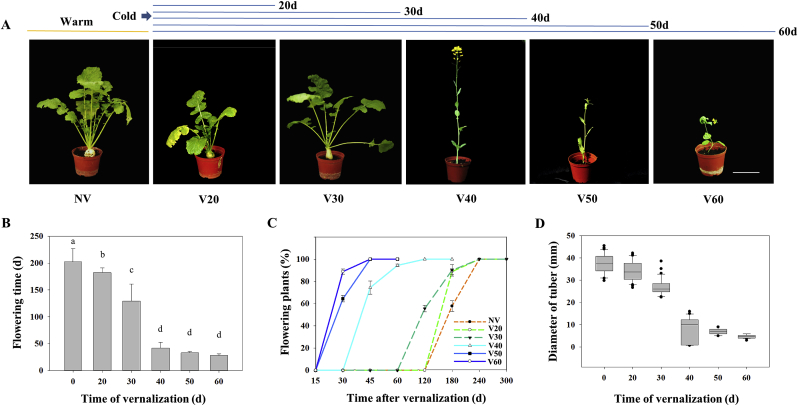
We firstly investigated the vernalization responses of Tibetan turnips in greenhouse experiments. Germinated seeds were subjected to cold treatment for 0–60 d. Non-vernalized turnips remained in the tuber growth phase for a long time and flowered very late, more than 200 d later (Fig. 1A–C). In contrast, prolonged vernalization was associated with flowering. After 40 d of vernalization, turnips flowered quickly upon being transferred to warm conditions for a short period of time. However, less than 30 days of vernalization was not sufficient to initiate flowering.
Fig. 1.
Phenotypes of Tibetan turnip in response to vernalization. A. Representative turnip plants cultivated without vernalization (NV) or with vernalization treatment (V20-60 d). Scale bar is 10 cm. B. Average flowering time (d, days to flower) after different vernalizaton treatments. C. Duration of the flowering phase after different times of vernalizaton. D. Diameter of tuber in response to cold exposure. Data are means ± SD of three biological replicates. ANOVA was performed for statistical analysis. Bars with different letters are significantly different from each other (P < 0.05).
We then measured tuber size after cold exposure. We found that tuber size decreased gradually over the course of vernalization (Fig. 1A and D). In the absence of vernalization, or partial vernalization (less than 30 d of cold), turnips produced obvious tuber characteristics. However, no tuber induction was observed when the plants were subjected to complete vernalization (more than 40 d of cold). These results suggest that vernalization effectively represses tuberization and promote flowering in Tibetan turnips.
3.2. Analysis of FLC homologues and their spatial expression in Tibetan turnips
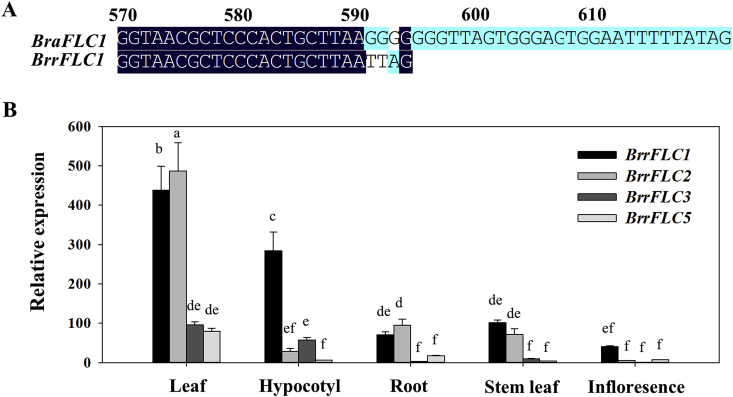
Previous studies have shown that non-vernalized plants and vernalized plants are significantly correlated to the expression of FLC in Arabidopsis (Shindo et al., 2005). To study the function of FLC homologues in the Tibetan turnip, we identified four paralogous FLC genes, namely, BrrFLC1, FLC2, FLC3, and FLC5. Because the turnip is a subspecies of B. rapa, we blasted these genes against the available Brassica A genome sequence from B. rapa (Cheng et al., 2011). Sequence alignments revealed that the same exon coding sequences in BrrFLC2, FLC3, and FLC5 were present in B. rapa. However, variants with different coding exons at the C-terminal were found in BrrFLC1 (Fig. 2A).
Fig. 2.
Identification and spatial expression analysis of BrrFLC homologues in Tibetan turnip. A. Comparison of FLC1 encoding sequences in C-terminal region between turnip and B. rapa. B. Spatial expression pattern of BrrFLC genes in different tissues of 20-day-old seedlings. Data are means ± SD of three biological replicates. ANOVA was performed for statistical analysis. Bars with different letters are significantly different from each other (P < 0.05).
We analyzed the spatial expression pattern of BrrFLC genes in different tissues using quantitative RT-PCR. Expression of BrrFLC genes was higher in leaves compared to all other tissues (Fig. 2B). In addition, the mRNA levels of the BrrFLC1 and BrrFLC2 transcripts were more than four times higher than those of BrrFLC3 and BrrFLC5 in leaves. BrrFLC1 was also highly expressed in the hypocotyl, whereas the other genes were not. Considering that the turnip tuber is primarily formed from the hypocotyl (Namikawa and Endo, 1932), the high transcript levels of BrrFLC1 in the hypocotyl might be associated with turnip development.
3.3. BrrFLC gene expression is related to tuberization and flowering during vernalization
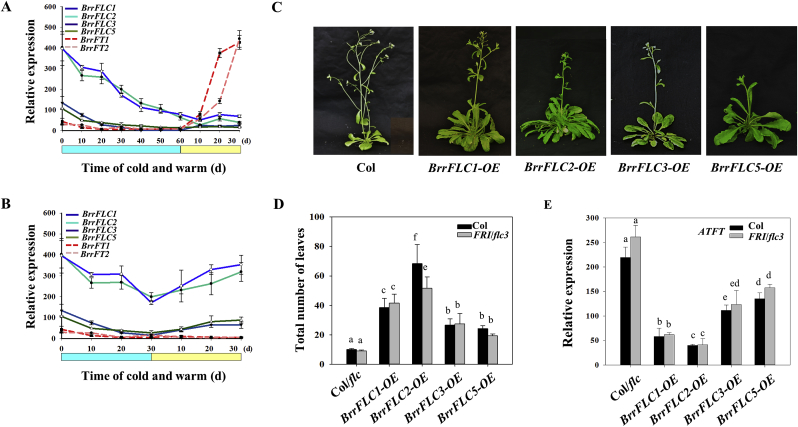
To examine the function of BrrFLC homologues of the Tibetan turnip in the vernalization response, we measured their expression following various cold treatments. BrrFLC1 and BrrFLC2 transcript levels decreased substantially in leaves during cold treatments, and eventually stabilized at low levels after 60 d, following the activation of the FT homologues BrrFT1 (reference to Bra022475) and BrrFT2 (reference to Bra004117) upon the transferral of the plants to warm conditions (Fig. 3A). The expression levels of BrrFLC3 and BrrFLC5 also decreased during vernalization, but not obviously so. Thirty days of vernalization was insufficient to completely silence BrrFLC gene expression and resulted in their reactivation after transferring plants to warm conditions, without BrrFT1 and BrrFT2 upregulation, and in tuberization and late flowering (Fig. 3B).
Fig. 3.
The major role of BrrFLC2 and BrrFLC1 in suppressing flowering. A. Expression analysis of BrrFLC genes in the leaves with 0–60 d of vernalization treatments. B. Expression analysis of BrrFLC genes in the leaves with 0–30 d of vernalization treatments. C. Flowering phenotypes of the transgenic lines of overexpression of BrrFLC genes in Arabidopsis. D. Flowering time of the transgenic lines in Col or Col-FRI-flc background. E. Quantitative RT–PCR analysis of the expression of AtFT in the leaves of transgenic lines and two wild-type strains. Two-week-old seedlings grown under LD conditions were used. Data are the means ± SD of three biological replicates. ANOVA was performed for statistical analysis. Bars with different letters are significantly different from each other (P < 0.05).
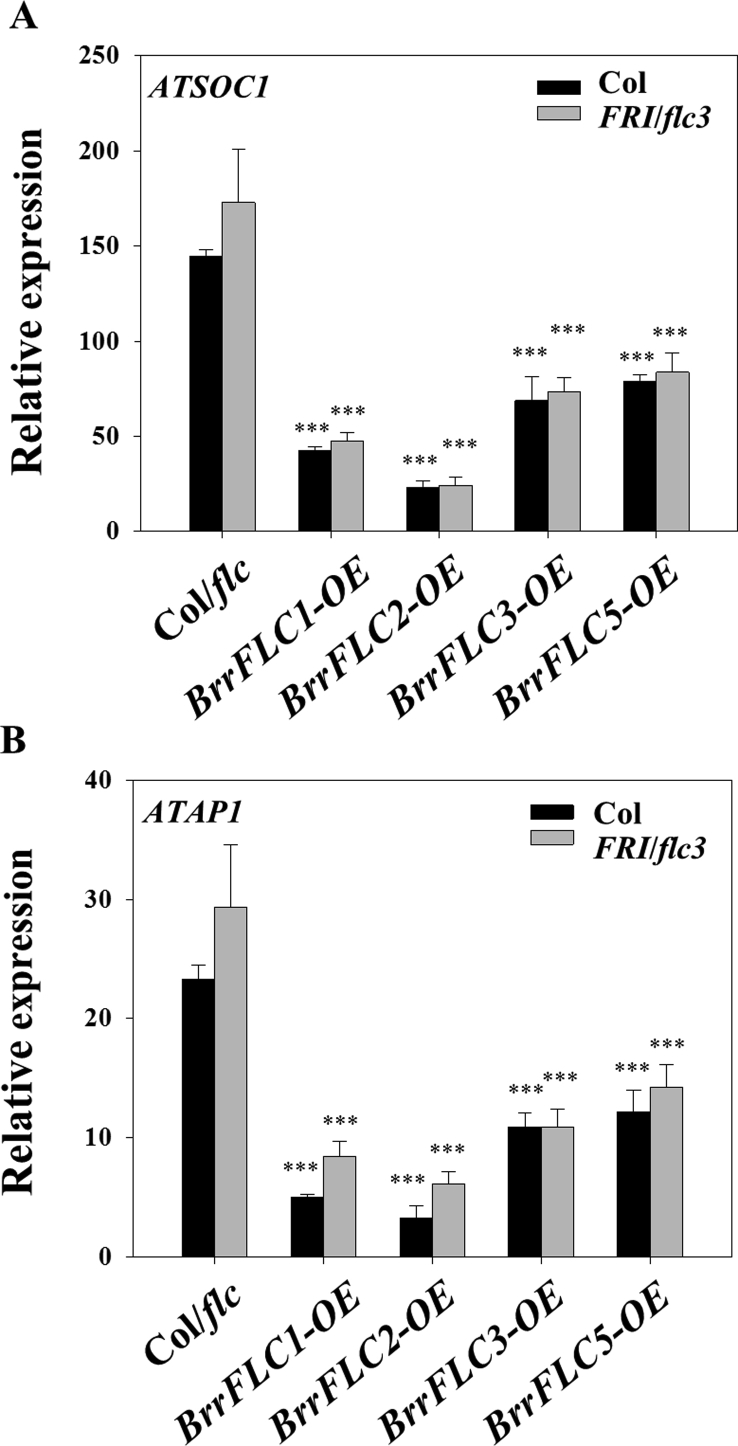
We then generated Arabidopsis transgenic lines overexpressing the BrrFLC genes in the Col and Col-FRI-flc backgrounds (Fig. S1). The overexpression of BrrFLC2 resulted in the greatest delays in flowering, followed by the BrrFLC1 overexpression line (Fig. 3C–D). Overexpression of BrrFLC3 and FLC5 also delayed flowering, but the effect was not as obvious as that of the BrrFLC2-OE and BrrFLC1-OE lines. These results indicate that the BrrFLC homologues in Tibetan turnips differentially contribute to the repression of flowering time. FLC is a transcriptional repressor that directly represses FT expression (Shindo et al., 2005). We therefore compared the expression levels of AtFT in transgenic and wild-type lines. As shown in Fig. 3E, the transcript levels of AtFT in the leaves of the transgenic lines were significantly lower than those in the two wild-type strains (P < 0.05). SOC1 and AP1 are marker genes for floral initiation. As observed in AtFT, AtSOC1 and AP1 expression was reduced in the scion apices of the transgenic lines (Fig. S2). Furthermore, the BrrFLC2-OE and BrrFLC1-OE lines exhibited lower AtFT transcript levels in the leaves, and lower AtSOC1 and AtAP1 levels in the scion apices than the BrrFLC3-OE and BrrFLC5-OE lines, which is consistent with the flowering phenotypes. Overall, these findings suggest that the expression of BrrFLC2 and BrrFLC1 in leaves plays a primary role in the suppression of flowering.
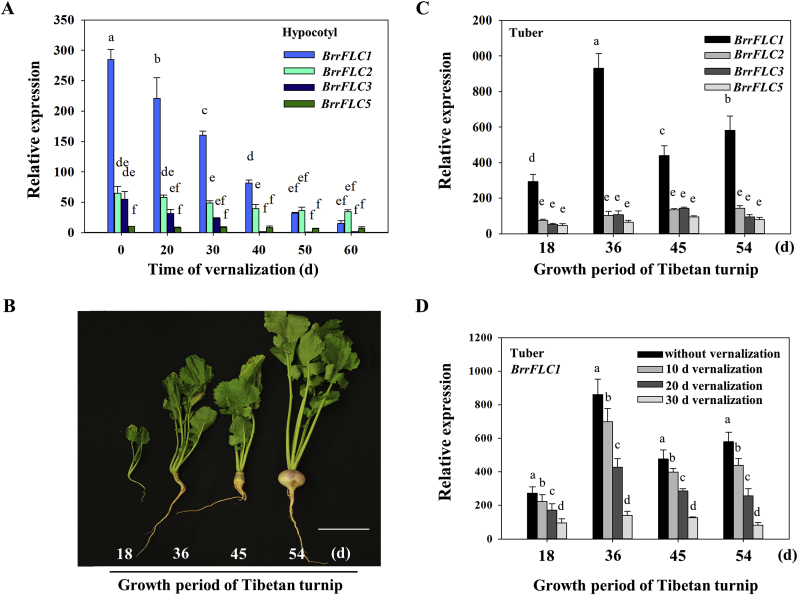
The high expression levels of BrrFLC1 in the hypocotyl was reduced sharply by cold treatment (Fig. 4A). Extended periods of cold treatment quantitatively regulated BrrFLC1 expression, ultimately resulting in its silencing. The expression of other BrrFLC genes was also influenced by vernalization, but the expression levels were very low. We further analyzed the expression levels of BrrFLC genes in the tuber of turnip plants at different growth periods. BrrFLC1 was moderately expressed in the tuber compared to the other BrrFLC genes. The highest expression level of BrrFLC1 was observed at 36 d (Fig. 4C), which corresponds to the critical stage of hypocotyl expansion (Fig. 4B). The vernalization treatment significantly reduced the transcriptional levels of BrrFLC1 in the tuber at different growth periods (Fig. 4D). These results indicated that the expression pattern of BrrFLC1 was associated with tuber enlargement and that BrrFLC1 responded to the vernalization treatment.
Fig. 4.
Expression analysis of BrrFLC genes in hypocotyl during vernalization and development. A. Expression analysis of BrrFLC genes in the hypocotyl during vernalization. B. Phenotypes of tuber formation in Tibetan turnip at different developmental stages (days after germination). C. Expression analysis of BrrFLC genes in the tuber of Tibetan turnip at different developmental stages. D. Expression analysis of BrrFLC genes in the tuber at different developmental stages after vernalization treatment. Data are means ± SD of three biological replicates. ANOVA was performed for statistical analysis. Bars with different letters are significantly different from each other (P < 0.05).
3.4. Grafting analysis of the role of vernalization on tuberization
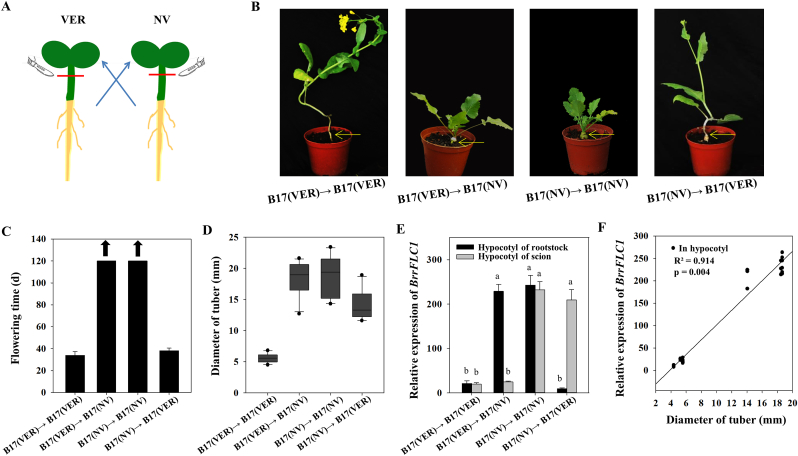
Grafting is an excellent tool for investigating molecular processes in plants. We thus established a hypocotyl micrografting system in the Tibetan turnip to further examine the role of vernalization on tuber formation (Fig. 5A). When the rootstock of the vernalized plant was grafted to the scion of the non-vernalized plant [termed B17(VER)→B17(NV)], the grafted line exhibited sustained vegetative growth with an obvious tuber in the scion above the grafting site (Fig. 5B–D). The expression level of BrrFLC1 in the tuber of the scion was high compared to the very low expression levels in the hypocotyl of the rootstock (Fig. 5E). For a control grafted line, we grafted the rootstock of a vernalized plant to the scion of a vernalized plant [termed B17(VER)→B17(VER)]. This line showed early flowering and no tuber formation; in addition, BrrFLC1 expression levels were low in the hypocotyl of both the scion and rootstock. We also created a B17(NV)→B17(VER) graft line which displayed early flowering in the scion and tuber formation in the rootstock (Fig. 5B–D). In the hypocotyl of the scion, BrrFLC1 expression levels were low, whereas in the tuber of the rootstock they were high (Fig. 5E). The control line B17(NV)→B17(NV) exhibited a similar phenotype to the B17(VER)→B17(NV) line, in which an obvious tuber had formed near the grafting site. Regression analyses also indicated a significant correlation between the expression levels of BrrFLC1 in the hypocotyl or tuber of the rootstock or scion and the tuber size in these graft lines (Fig. 5F). These results indicate that vernalization effectively suppresses tuberization in the hypocotyl of the rootstock or scion independently, and that BrrFLC1 expression is correlated with this process.
Fig. 5.
Phenotype analysis and expression analysis of BrrFLC1 in grafted plants with or without vernalization. A. Sketch map of grafting in Tibetan turnip. B. Phenotypes of various grafted plants between non-vernalized (NV) and vernalized (VER) turnip. C. Flowering time (d) of different grafted plants. Up arrow represents no flowering at the end of the experiment after 120 d of growth. D. Diameter of tuber of different grafted plants. E. Expression analysis of BrrFLC1 in the hypocotyl of both the scion and rootstock in grafted plants. F. Relationship between tuber size and BrrFLC1 expression in grafted plants. Data are means ± SD of three biological replicates. ANOVA was performed for statistical analysis. Bars with different letters are significantly different from each other (P < 0.05). R2 and p depict the square of the Pearson correlation coefficient and its associated p value, respectively. The solid lines represent linear regression trend lines.
4. Discussion
In this study, we functionally characterized four FLC homologues (BrrFLC1, FLC2, FLC3, and FLC5) in the Tibetan turnip and found that BrrFLC2 and BrrFLC1 were significantly associated with vernalization response in a quantitative way. Plants with a vernalization requirement need varying levels of cold exposure to initiate flowering (Yan et al., 2004). It has been shown that some species and ecotypes from high altitudes require a strict vernalization requirement (Méndezvigo et al., 2011). Our results show that at least a 40-day vernalization treatment is required to initiate flowering in the Tibetan turnip. The correlation between the expression levels of BrrFLC genes and flowering time and the genetic analyses in Arabidopsis transgenic lines indicate that BrrFLC2 and BrrFLC1 act as the main repressors of flowering and determine the length of the vernalization response.
In addition, we discovered high BrrFLC1 transcript levels in the hypocotyl and tuber. It has been previously shown that the fleshy tubers of turnips are formed mainly through secondary growth of the xylem in the hypocotyl (Namikawa and Endo, 1932). In our study, BrrFLC1 expression peaked at 36 d, which constitutes the critical period of tuber enlargement. Cold treatment may also effectively repress BrrFLC1 expression in the hypocotyl, resulting in the inhibition of tuberization. Previous studies have shown that in addition to its role in repressing flowering, FLC may be involved in other developmental pathways by binding promoters of many genes in Arabidopsis (Deng et al., 2011). The correlation between BrrFLC1 expression and tuber expansion suggest that BrrFLC1 acts as an indicator in tuber induction (Fig. 2).
The majority of research on tuber formation to date has focused on widely cultivated tuber crops, such as potato (Solanum tuberosum) (Kloosterman et al., 2005), sugar beet (Beta vulgaris) (Lukaszewska et al., 2012), and radish (Raphanus sativus) (Xu et al., 2013), and has provided insight into the molecular mechanisms of tuberization. To further investigate the role of vernalization on tuber formation, we established a hypocotyl micrografting system in the Tibetan turnip. Using grafting approaches, deep molecular knowledge has been obtained in different processes (Ayre and Turgeon, 2004, Buhtz et al., 2010, Liang et al., 2012). Due to the nascent state of the turnip transformation system, the grafting method offers a suitable alternative for studying the relationship between vernalization, tuber size, and BrrFLC gene expression. Diverse tuber sizes and flowering times were identified in the graft lines, and were significantly correlated with BrrFLC1 expression in the rootstock or scion independently. In summary, we identified high BrrFLC1 transcript levels at a particular stage of tuber development in the tuber tissue that are associated with the function of vernalization on tuberization. Further studies will focus on the contribution of BrrFLC homologues to different development pathways, using molecular tools such as quantitative trait loci (QTLs) analyses and high-throughput sequencing of ChIP-seq analyses.
Acknowledgements
This work was supported by the National Science Foundation of China (No. 31500221, 31590823 and 31601999) and the West Light Foundation of the Chinese Academy of Sciences by XXK.
(Editor: Gang Liang)
Footnotes
Peer review under responsibility of Editorial Office of Plant Diversity.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.pld.2018.01.002.
Contributor Information
Xiangxiang Kong, Email: kongxiangxiang@mail.kib.ac.cn.
Yongping Yang, Email: yangyp@mail.kib.ac.cn.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
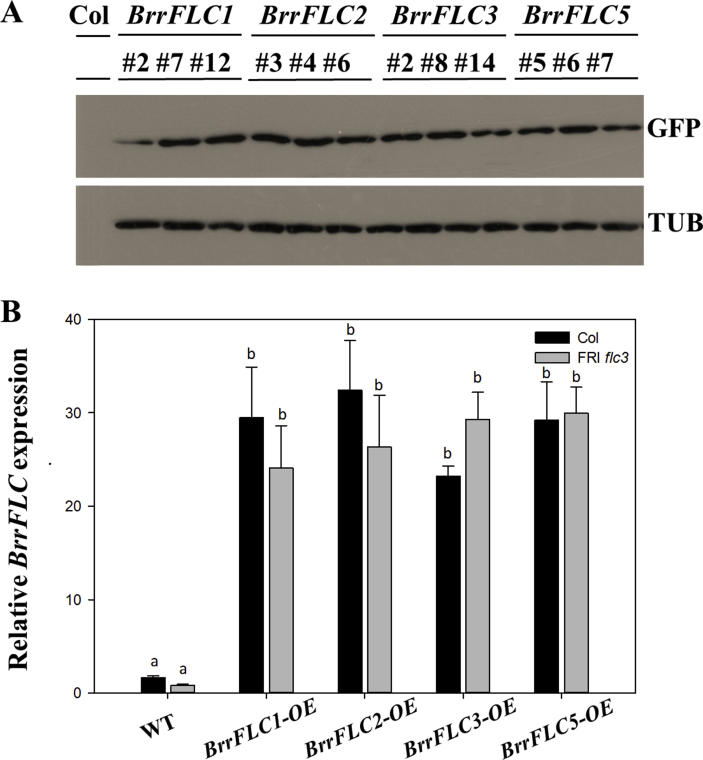
Supplemental Fig. 1.
Identification of the transgenic lines. A. Immunoblot analysis of BrrFLC gene expression, each using anti-GFP antibody. Protein extracted from the wild-type Col line and three individual transgenic lines respectively. B. BrrFLC expression in transgenic lines and ATFLC expression in two wild-type strains. Two-week-old seedlings grown under LD conditions were used. Data are means ± SD of three biological replicates. ANOVA was performed for statistical analysis. Bars with different letters are significantly different from each other (P < 0.05).
Supplemental Fig. 2.
Quantitative RT–PCR analysis of the expression of AtAP1 and AtSOC1 in the shoot apices of transgenic lines and two wild-type strains. Two-week-old seedlings grown under LD conditions were used. Data are the means ± SD of three biological replicates. Statistical analyses were performed using a t test. ***, p < 0.001; **, p < 0.01; and *, p < 0.05.
Primers used in this study.
References
- Ayre B.G., Turgeon R. Graft transmission of a floral stimulant derived from CONSTANS. Plant Physiol. 2004;135:2271–2278. doi: 10.1104/pp.104.040592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastow R., Mylne J.S., Lister C., Lippman Z., Martienssen R.A., Dean C. Vernalization requires epigenetic silencing of FLC by histone methylation. Nature. 2004;427:164. doi: 10.1038/nature02269. [DOI] [PubMed] [Google Scholar]
- Buhtz A., Pieritz J., Springer F., Kehr J. Phloem small RNAs, nutrient stress responses, and systemic mobility. BMC Plant Biol. 2010;10:64. doi: 10.1186/1471-2229-10-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F., Liu S., Wu J., Fang L., Sun S., Liu B., Li P., Hua W., Wang X. BRAD, the genetics and genomics database for Brassica plants. BMC Plant Biol. 2011;11:136. doi: 10.1186/1471-2229-11-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu B., Chen C., Li J., Chen X., Li Y., Tang W., Jin L., Zhang Y. Effects of Tibetan turnip (Brassica rapa L.) on promoting hypoxia-tolerance in healthy humans. J. Ethnopharmacol. 2017;195:246–254. doi: 10.1016/j.jep.2016.11.028. [DOI] [PubMed] [Google Scholar]
- De Lucia F., Crevillen P., Jones A.M., Greb T., Dean C. A PHD-polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc. Natl. Acad. Sci. U. S. A. 2008;105:16831–16836. doi: 10.1073/pnas.0808687105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., Ying H., Helliwell C.A., Taylor J.M., Peacock W.J., Dennis E.S. FLOWERING LOCUS C (FLC) regulates development pathways throughout the life cycle of Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 2011;108:6680–6685. doi: 10.1073/pnas.1103175108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J., Yan L., Raman H., Zou X., Jing W., Dai S., Xiao Q., Cong L., Fan L., Liu B. A Tourist -like MITE insertion in the upstream region of the BnFLC.A10 gene is associated with vernalization requirement in rapeseed ( Brassica napus L. ) BMC Plant Biol. 2012;12:238. doi: 10.1186/1471-2229-12-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin J.A., Soumpourou E., Lister C., Ligthart J.D., Kennedy S., Dean C. Nucleotide polymorphism affecting FLC expression underpins heading date variation in horticultural brassicas. Plant J. 2016;87:597. doi: 10.1111/tpj.13221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.H., Doyle M.R., Sung S., Amasino R.M. Vernalization: winter and the timing of flowering in plants. Annu. Rev. Cell Dev. Biol. 2009;25:277. doi: 10.1146/annurev.cellbio.042308.113411. [DOI] [PubMed] [Google Scholar]
- Kloosterman B., Vorst O., Hall R.D., Visser R.G., Bachem C.W. Tuber on a chip: differential gene expression during potato tuber development. Plant Biochem. J. 2005;3:505–519. doi: 10.1111/j.1467-7652.2005.00141.x. [DOI] [PubMed] [Google Scholar]
- Krichevsky A., Gutgarts H., Kozlovsky S.V., Tzfira T., Sutton A., Sternglanz R., Mandel G., Citovsky V. C2H2 zinc finger-SET histone methyltransferase is a plant-specific chromatin modifier. Dev. Biol. 2007;303:259–269. doi: 10.1016/j.ydbio.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D., White R.G., Waterhouse P.M. Gene silencing in Arabidopsis spreads from the root to the shoot, through a gating barrier, by template-dependent, nonvascular, cell-to-cell movement. Plant Physiol. 2012;159:984–1000. doi: 10.1104/pp.112.197129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y.S., Kim H.K., Lefeber A.W., Erkelens C., Choi Y.H., Verpoorte R. Identification of phenylpropanoids in methyl jasmonate treated Brassica rapa leaves using two-dimensional nuclear magnetic resonance spectroscopy. J. Chromatogr. A. 2006;1112:148–155. doi: 10.1016/j.chroma.2005.11.114. [DOI] [PubMed] [Google Scholar]
- Lukaszewska E., Virden R., Sliwinska E. Hormonal control of endoreduplication in sugar beet (Beta vulgaris L.) seedlings growing in vitro. Plant Biol. 2012;14:216–222. doi: 10.1111/j.1438-8677.2011.00477.x. [DOI] [PubMed] [Google Scholar]
- Méndezvigo B., Picó F.X., Ramiro M., Martínezzapater J.M., Alonsoblanco C. Altitudinal and climatic adaptation is mediated by flowering traits and FRI, FLC, and PHYC genes in Arabidopsis. Plant Physiol. 2011;157:1942–1955. doi: 10.1104/pp.111.183426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsch-Martínez N., Franken J., Gonzalez-Aguilera K.L., de Folter S., Angenent G., Alvarez-Buylla E.R. An efficient flat-surface collar-free grafting method for Arabidopsis thaliana seedlings. Plant Meth. 2013;9:14. doi: 10.1186/1746-4811-9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S.D., Himelblau E., Kim S.Y., Schomburg F.M., Amasino R.M. Integration of flowering signals in winter-annual Arabidopsis. Plant Physiol. 2005;137:149–156. doi: 10.1104/pp.104.052811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namikawa K., Endo M. Anatomical analysis of root growth of turnip cv. Sugukina (Brassica rapa L. Agric. Hort. (Nogyo Oyobi Engei) 1932;7:3–16. [Google Scholar]
- Okazaki K., Sakamoto K., Kikuchi R., Saito A., Togashi E., Kuginuki Y., Matsumoto S., Hirai M. Mapping and characterization of FLC homologs and QTL analysis of flowering time in Brassica oleracea. Theor. Appl. Genet. 2007;114:595–608. doi: 10.1007/s00122-006-0460-6. [DOI] [PubMed] [Google Scholar]
- Ruelens P., de Maagd R.A., Proost S., Theißen G., Geuten K., Kaufmann K. FLOWERING LOCUS C in monocots and the tandem origin of angiosperm-specific MADS-box genes. Nat. Commun. 2013;4:2280. doi: 10.1038/ncomms3280. [DOI] [PubMed] [Google Scholar]
- Schmitz R.J., Amasino R.M. Vernalization: a model for investigating epigenetics and eukaryotic gene regulation in plants. Biochim. Biophys. Acta. 2007;1769:269–275. doi: 10.1016/j.bbaexp.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Schranz M.E., Quijada P., Sung S.B., Lukens L., Amasino R., Osborn T.C. Characterization and effects of the replicated flowering time gene FLC in Brassica rapa. Genetics. 2002;162:1457. doi: 10.1093/genetics/162.3.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N., Ruelens P., D'Hauw M., Maggen T., Dochy N., Torfs S., Kaufmann K., Rohde A., Geuten K. A flowering locus C homolog is a vernalization-regulated repressor in brachypodium and is cold regulated in wheat. Plant Physiol. 2017;173:1301–1315. doi: 10.1104/pp.16.01161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo C., Aranzana M.J., Lister C., Baxter C., Nicholls C., Nordborg M., Dean C. Role of FRIGIDA and FLOWERING LOCUS C in determining variation in flowering time of Arabidopsis. Plant Physiol. 2005;138:1163–1173. doi: 10.1104/pp.105.061309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Zhu X., Chen Y., Gong Y., Liu L. Expression profiling of genes involved in ascorbate biosynthesis and recycling during fleshy root development in radish. Plant Physiol. Biochem. 2013;70:269–277. doi: 10.1016/j.plaphy.2013.05.041. [DOI] [PubMed] [Google Scholar]
- Yan L., Loukoianov A., Blechl A., Tranquilli G., Ramakrishna W., SanMiguel P., Bennetzen J.L., Echenique V., Dubcovsky J. The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science. 2004;303:1640–1644. doi: 10.1126/science.1094305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y.X., Wu J., Sun R.F., Zhang X.W., Xu D.H., Bonnema G., Wang X.W. A naturally occurring splicing site mutation in the Brassica rapa FLC1 gene is associated with variation in flowering time. J. Exp. Bot. 2009;60:1299. doi: 10.1093/jxb/erp010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Kulkarni V., Liu N., Del Carpio D.P., Bucher J., Bonnema G. BrFLC2 (FLOWERING LOCUS C) as a candidate gene for a vernalization response QTL in Brassica rapa. J. Exp. Bot. 2010;61:1817–1825. doi: 10.1093/jxb/erq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used in this study.