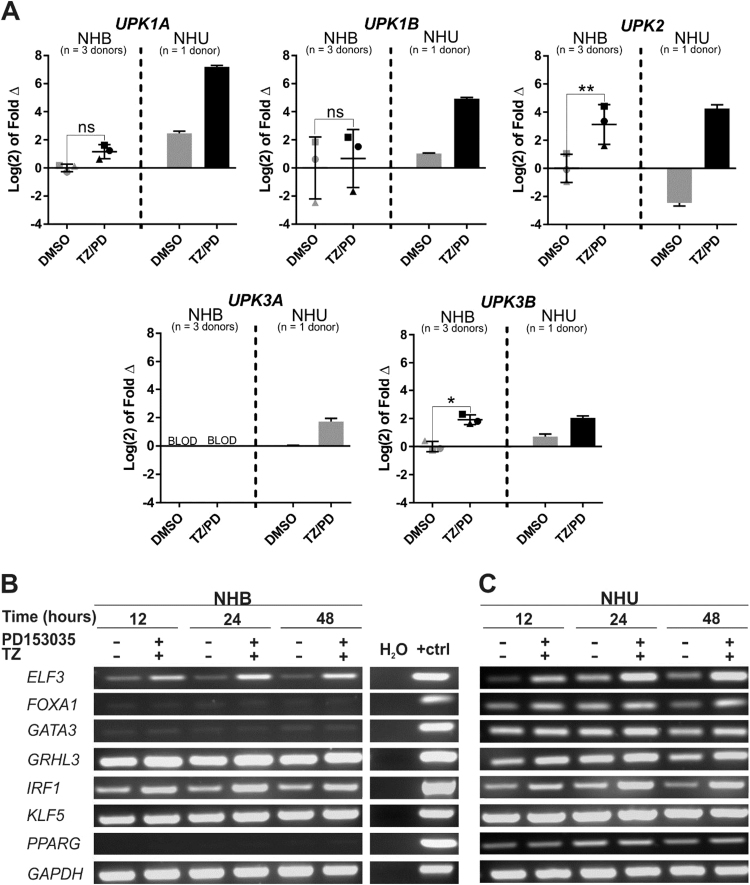
Fig. 3.
Comparison of uroplakin and urothelium differentiation-associated transcription factor gene expression by NHB and NHU cell cultures. Employing protocols developed to differentiate NHU cells by PPARγ activation, cell cultures of NHB or NHU cells were exposed to 1 µM troglitazone and 1 µM PD153035 (TZ/PD) for 24 h, maintained in 1 µM PD153035 and harvested at 12, 24, 48 and/or 72 h. Control cultures were exposed to vehicle (0.1% DMSO) alone. (A) RTqPCR for three independent NHB cell lines (represented by different symbols), versus a single NHU cell line for comparison of uroplakin (UPK1A, UPK1B, UPK2, UPK3A and UPK3B) mRNA expression at the 72 h time-point. All data has been normalised to GAPDH expression and is presented relative to the DMSO-treated NHB cells for each gene except UPK3A, where the data is shown relative to the DMSO treated NHU cells due to absent UPK3A gene expression by NHB cells. BLOD = Below Limit of Detection. Statistical analysis was performed using a two-tailed, paired t-test to determine whether TZ/PD resulted in any significant change in gene expression in NHB cells. *represents P ≤ 0.05, ** represents P ≤ 0.01. Error bars represent standard deviation. (B-C) RT-PCR of ELF3, FOXA1, GATA3, GRHL3, IRF1, KLF5 and PPARG mRNA expression by (B) NHB cells and (C) NHU cells. RNA was extracted at the 12, 24 and 48 h time-points and then DNAase-treated and used to generate cDNA for RT-PCR. GAPDH was used as an internal loading control. A no-template (H2O) control was included as a negative control for the PCR reaction, and genomic DNA was used as the positive control (+ctrl). No product was amplified from RT-negative controls (not shown). Experiments were performed on n = 2 independent NHB donor cell lines and representative results shown.