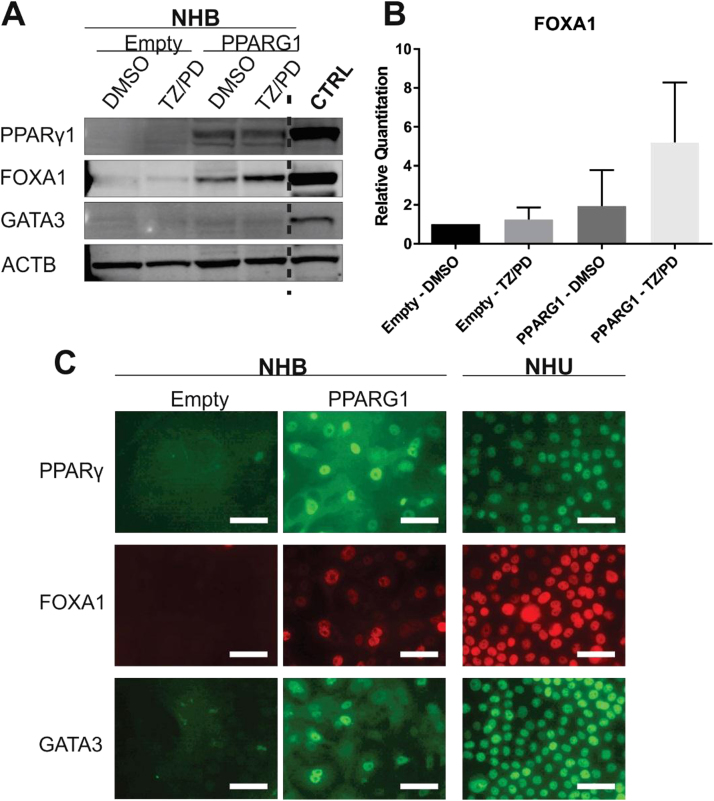
Fig. 6.
Evaluation of PPARγ, FOXA1 and GATA3 expression in PPARG1 overexpressing and control (empty) NHB cells. PPARG1 overexpressing and control (empty vector) NHB cell cultures were exposed to the TZ/PD protocol for 72 h. Experiments performed on between 2 and 4 independent NHB cell lines (as stated below), with representative results shown. (A) PPARγ1, FOXA1 and GATA3 protein expression assessed by western blotting. ACTB expression was included as an internal loading control. Protein lysates from cell lines known to express the proteins of interest were included as positive controls for each antibody (CTRL). Experiments performed on n = 3 independent NHB cell lines. (B) Densitometry analysis of FOXA1 protein expression shown relative to control (Empty - DMSO) NHB cells. Data is shown as the mean of n = 4 independent transduced NHB cell lines. All values were normalised to the ACTB expression. Statistical analysis was performed using a one-way ANOVA test, but no statistical significance was found (P > 0.05). Error bars represent standard deviation. (C) Immunofluorescence microscopy evaluating PPARγ, FOXA1 and GATA3 protein localisation in PPARG1 overexpressing and control (empty vector) NHB cells following the TZ/PD protocol at 72 h. Experiments were performed on n = 3 independent transduced NHB cell lines. IF images for a single NHB cell line are shown. Nuclear localisation was observed with PPARγ (n = 3/3), FOXA1 (n = 3/3), and GATA3 (n = 2/3). NHU cells (non-transduced) treated with the TZ/PD protocol are shown for comparison at the same time point. Scale bar ≡ 50 µm.