Figure 6: Single molecule load-dependent kinetics as the basis of the ensemble force-velocity relationship in β-cardiac myosin.
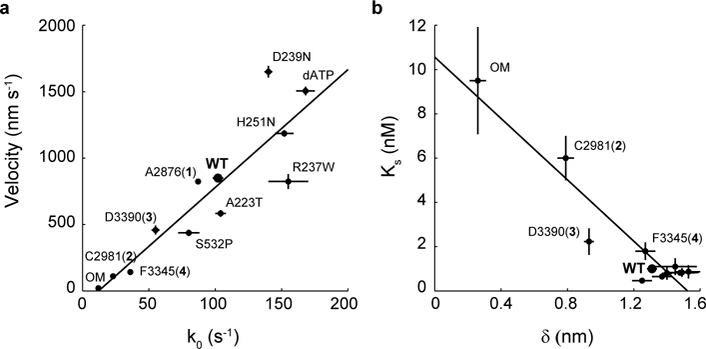
a, Unloaded actin sliding velocity measured by in vitro motility assay vs. k0, the detachment rate at zero load for a single myosin molecule, for the compounds and mutations studied. Linear regression gives a slope of 8.9 nm and R2 = 0.81. Note that some points have error bars smaller than the dot. b, The parameter Ks from the loaded motility assay is a measure of load-bearing ability at the ensemble level, while 𝛿 is the load-sensitivity parameter determined at the single molecule level. Pearson correlation −0.94. For clarity, the cluster of data points at the lower right corner are not individually labeled. Motility velocity and Ks values of mutants and dATP are obtained from other studies 20,25,32,44 (Tomasic I., Liu C., Rodriguez H., Spudich J.A., Bartholomew Ingle S.R., manuscript in preparation). Error bars represent s.e.m.
