Figure S1.
Transcriptome Analysis: Correcting for Loss of Multimapping Loss, Related to Figure 1
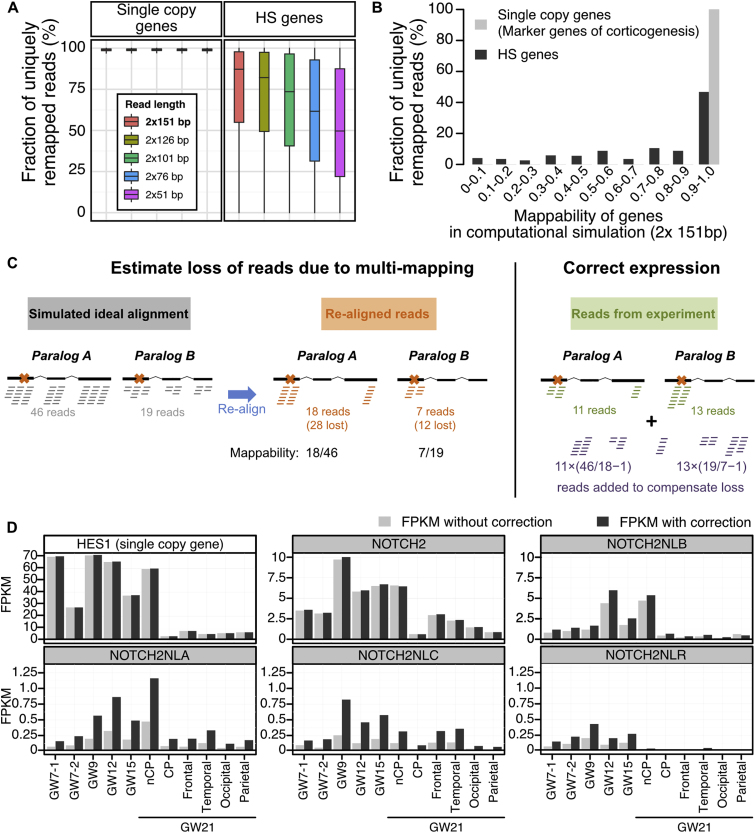
(A) The mappability of a gene is defined as the number of reads uniquely mapped on this gene divided by the number of reads originating from its transcripts. It is computed from simulated transcriptomes, as explained above and depicted in panel C. Most reads originating from single copy genes are uniquely mapped on the reference genome regardless of sequencing length, hence they have a mappability of one. HS genes show lower mappability values. As expected, their mappability is inversely related to read length.
(B) When setting simulated read length to 2x 151bp, which is the length used for transcriptome sequencing in this study, only half of the HS genes show mappability values above 90%.
(C) Computational correction of expression for human specific paralogs. Paralogs within each HS gene families are highly similar; potentially confusing the mapping of reads originating from individual paralogs. As a result, some reads are discarded because they map to multiple paralogs, leading to expression under-estimation. To estimate this loss quantitatively, an alignment of simulated reads (BAM file) is generated for each gene (gray alignment) at a defined coverage (see methods). This simulated alignment is ideal as it assumes a uniform coverage of the genes, and importantly the reads are manufactured and placed on reference genome, i.e., no read mapping procedure is involved, hence there is no mapping ambiguity. These simulated reads are then extracted and aligned with the same alignment procedure as used for in vivo experimental data (see methods; orange alignment, crosses on the gene structures denote unique sequence features allowing unambiguous mapping). Many reads are lost in the process due to multimapping, but we can estimate how many, since we initially generated them in known quantity (i.e., gray alignment). Finally, when aligning reads from in vivo experiments (green alignment), these estimates are used to inject in the alignments the near-exact number of additional reads to compensate for the loss of multimapping reads (purple alignment).
(D) Example of correction: FPKM values computed without (light gray) and with the simulation-based correction (dark gray) for 5 paralogous genes of the NOTCH2 family and HES1 as an example of single copy gene.