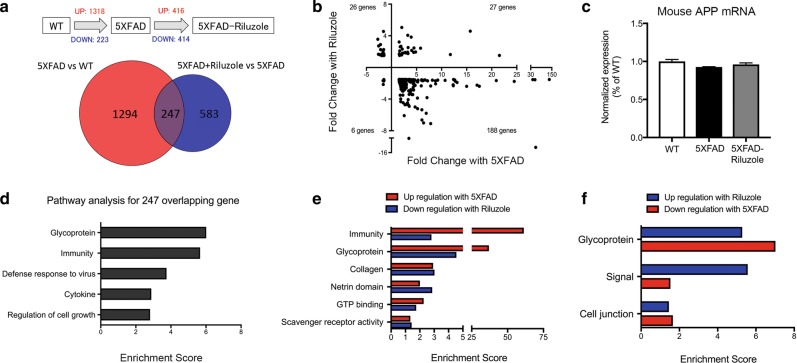
Fig. 3. Treatment of 5XFAD mice with riluzole rescues gene expression changes in the hippocampus.
a Venn diagram illustrating the overlap of 247 genes that were changed in 5XFAD compared to wild type and in 5XFAD mice treated with riluzole compare to untreated mice. b Scatter plot illustrating the 247 overlapping genes showing fold change by 5XFAD (x-axis) against fold change with riluzole treatment on 5XFAD mice (y-axis). 86% of overlapping 5XFAD genes is reversed by riluzole treatment. The upper left quadrant represents 26 genes that had decreased expression with 5XFAD and increased expression after riluzole treatment. Conversely, the lower-right quadrant illustrates 189 genes that were increased with 5XFAD and decreased by riluzole. c The mRNA expression of Mouse APP from RNA-Seq data revealed no significant difference among WT, 5XFAD, and 5XFAD-Riluzole groups. d Histograms illustrating significantly enriched pathways based on genes differentially expressed by overlapping gene. (enrichment score > 1.3). e, f Histograms illustrating significantly enriched pathways with the highest sum enrichment scores across opposite conditions (Increased in 5XFAD and Decreased with riluzole treatment or vice-versa) based on differentially expressed genes (enrichment score > 1.3). Similar pathways and enrichment scores were observed when comparing genes decreased by 5XFAD and increased by riluzole, as well as for genes increased with age and decreased by riluzole. The RNA-Seq data is based on hippocampal tissue pooled into three replicate sequencing libraries/group from WT, n = 10 (pooled 3, 3, 4 mice); 5XFAD, n = 7 (pooled 2, 2, 3 mice); and 5XFAD-Riluzole, n = 8 (pooled 2, 3, 3 mice)