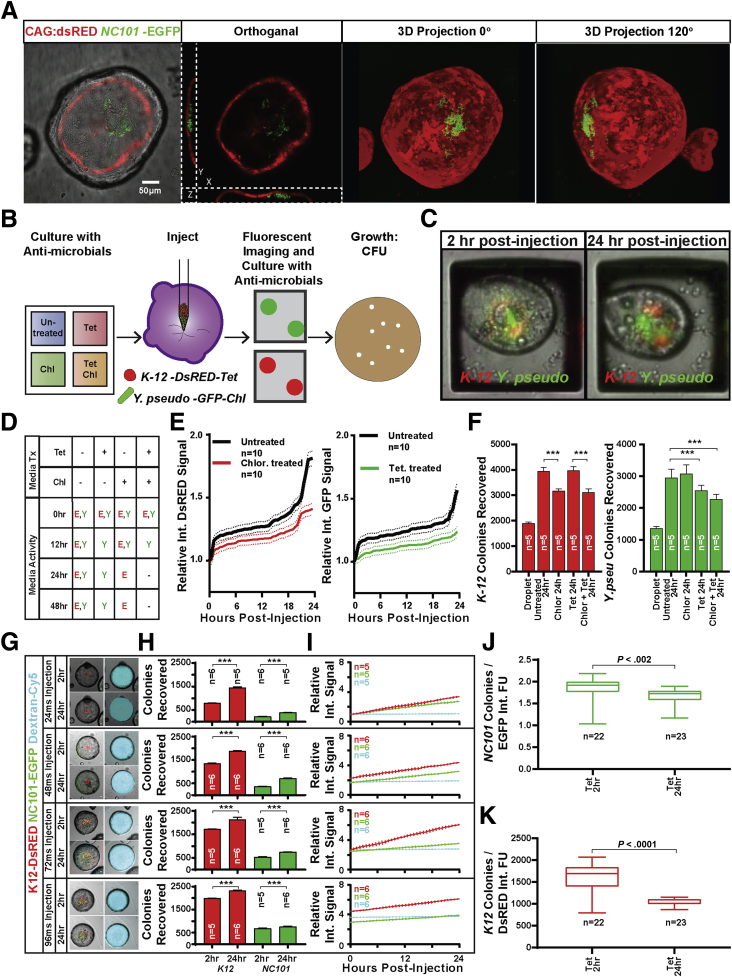
Figure 3.
The colonoid lumen forms a discrete compartment compatible with specific microbial growth. (A) GFP-expressing E coli can be visualized after microinjection into DsRED-expressing colonoids and appears to sit in the bottom of the lumen cavity in all colonoids observed. (B) The effects of antibiotics in the colonoid culture media on lumen microbe compatibility was investigated using a CRA device to culture colonoids in 4 discrete reservoirs treated with tetracycline and/or chloramphenicol. Colonoids from each well were targeted for microinjection with a mixed microbial culture of DsRED-expressing E coli resistant to tetracycline and GFP-expressing Y pseudotuberculosis resistant to chloramphenicol. Injected colonoids were monitored over time by live fluorescent imaging before lumen contents were harvested to assess microbial growth by colony formation on conventional agar plates. (C) Fluorescent signal from both microbes could be observed within the lumen of successfully injected colonoids during the entire time course. (D) Antibiotics were essential for preventing off-target growth by excess bacteria delivered to the media during microinjection with no active microbes discovered in culture media treated with chloramphenicol and tetracycline 24 hours after microinjection. (E) Computational analysis showed an increase in integrated DsRED and EGFP fluorescence signal of raft images containing successfully injected colonoids, suggesting an increase in DsRED- and EGFP-expressing microbes. (F) More E coli and Y pseudotuberculosis colonies were recovered from the lumen of colonoids from all media conditions compared with the input injection droplet, suggesting the colonoid lumen protected the injected microbes from chloramphenicol and tetracycline delivered in the culture media (n = 10 colonoids in each condition). Significantly more colonies were recovered from untreated colonoids, correlating with increased integrated fluorescence signal. (G) Fluorescent signal from both microbes as well as inert fluorescent cargo could be observed within the lumen of successfully injected colonoids during the entire time course. (H) More K12 and NC101 colonies were recovered from colonoids collected 24 hours after microinjection compared with those collected immediately after microinjection, suggesting that both microbes grew regardless of the delivered load (n = 5–6 colonoids from each injection duration). (I) Computational analysis showed an increase in integrated DsRED and EGFP fluorescence signal in injected colonoids, suggesting an increase in DsRED- and EGFP-expressing microbes. Computational analysis also showed stable integrated Alexa Fluor 647 signal, suggesting that delivered dextran was well retained. (J) The measured ratio of recovered NC101 colonies to integrated EGFP signal varied significantly between the 2-hour and 24-hour time points, suggesting that integrated EGFP signal cannot be used to directly measure E coli NC101-EGFP microbial load. (K) The measured ratio of recovered K12 colonies to integrated EGFP signal varied significantly between the 2-hour and 24-hour time points, suggesting that integrated EGFP signal cannot be used to directly measure E coli–DsRED microbial load. Chl, chloramphenicol; FU, follow-up evaluation; Int, integrated; Tet, tetracycline; Tx, treatment; Y pseudo, Y pseudotuberculosis.