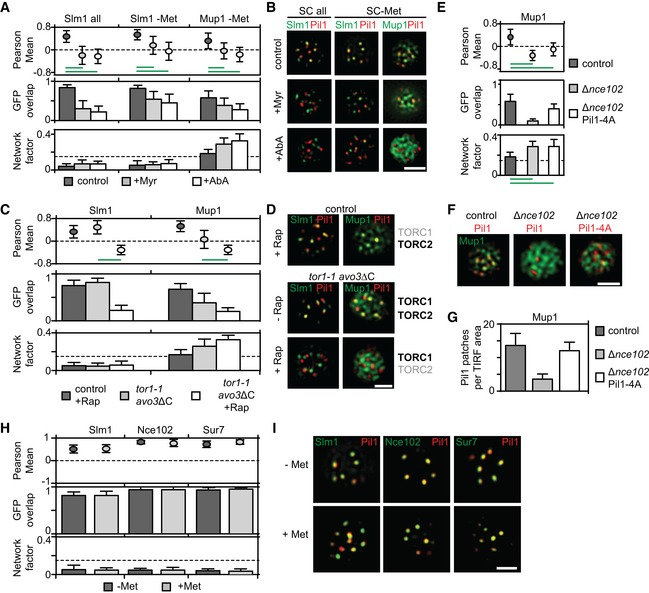
Effect of sphingolipid stress on relocation of Slm1 and on Mup1 clustering in the absence of Met. Colocalization with Pil1‐RFP (Pearson Mean), GFP overlap (fraction of the GFP signal found within the structure marked by Pil1‐RFP), and Network factor are given. Myriocin (Myr) blocks the first step in sphingolipid synthesis, while aureobasidin A (AbA) blocks the formation of complex sphingolipids.
Representative two‐color TIRFM images from the experiments summarized in (A).
Degrees of Mup1 clustering in the MCC in the indicated strains and upon TORC1/2 inhibition with rapamycin (Rap). The tor1‐1 mutant makes TORC1 rapamycin‐insensitive, while the avo3ΔC mutant renders TORC2 rapamycin‐sensitive.
Representative two‐color TIRFM images from the experiments summarized in (C). Light gray labels indicate inactive TOR complexes; bold labels indicate active TOR complexes.
Nce102 is required for Mup1 clustering into the MCC.
Representative two‐color TIRFM images of the Nce102‐dependent clustering of Mup1‐GFP in the PM from the experiments summarized in (E).
Nce102‐dependent eisosome stability shown as number of Pil1‐RFP or Pil1‐4A‐RFP patches per area.
Lateral segregation of Slm1, Nce102, and Sur7 before and after Met addition.
Representative two‐color TIRFM images from the experiments summarized in (H).
= 50–300 cells. Green lines indicate significantly different data sets. An overview of the performed statistics can be found in
. Scale bars: 2 μm. All values are listed in
.