Figure 2.
Rapid and High-Efficiency Derivation of MSCs
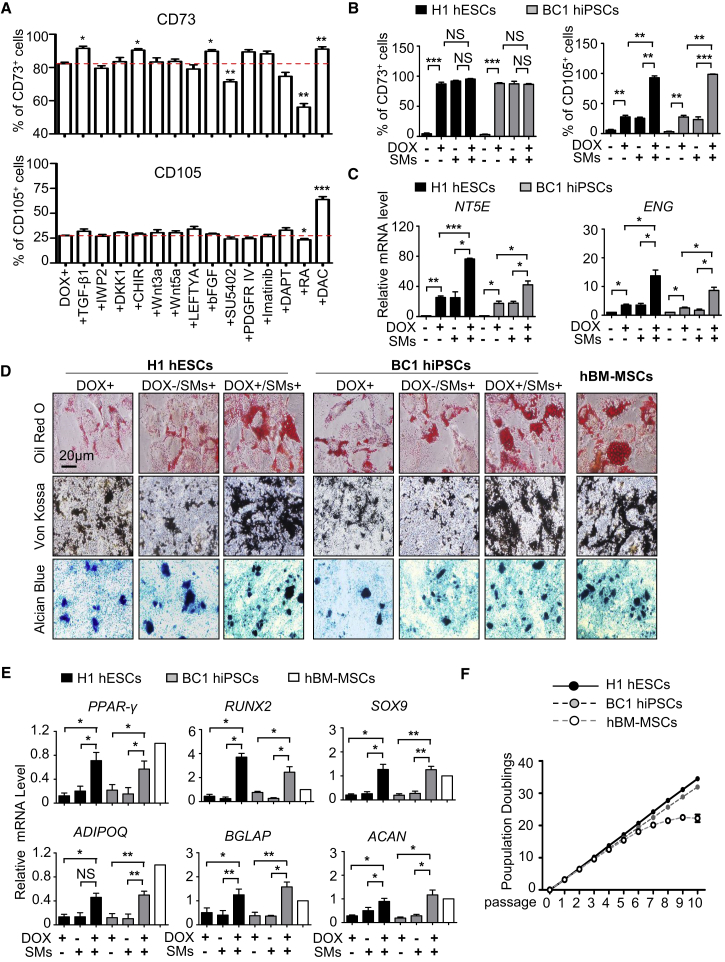
(A) FCM analysis for MSC markers of GFP-MSX2 H1 hESCs with 3 μg/mL DOX alone or indicated chemical compound addition (see Table S3) for 7 days (mean ± SEM, N = 3). ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
(B) FCM analysis of MSC markers in GFP-MSX2 hPSCs (H1, BC1) with indicated treatments for 7 days (mean ± SEM, N = 3). ∗∗p < 0.01; ∗∗∗p < 0.001; NS, not significant.
(C) qRT-PCR analysis of MSC markers in GFP-MSX2 hPSCs (H1, BC1) with indicated treatments for 7 days (mean ± SEM, N = 3). ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. Values are normalized to the Dox− group (=1).
(D) Tri-lineage differentiation potential of MC-MSCs derived from GFP-MSX2 hPSCs (H1, BC1) and hBM-MSCs for the indicated lineages. Scale bar, 20 μm.
(E) Relative expression levels of genes associated with tri-lineage differentiation of the MC-MSCs derived from GFP-MSX2 hPSCs (H1, BC1) (mean ± SEM, N = 3). ∗p < 0.05; ∗∗p < 0.01. Values are normalized to the hBM-MSCs group (=1).
(F) Expansion potential of MC-MSCs derived from GFP-MSX2 hPSCs (H1, BC1) and hBM-MSCs in MSC culture media with CHIR99021 (0.5 μM) for 10 passages by population doubling assay (mean ± SEM, N = 3).
See also Figure S2.