Figure 4.
Neural Crest as the Intermediate Stage between Pluripotency and Mesenchymal Fate
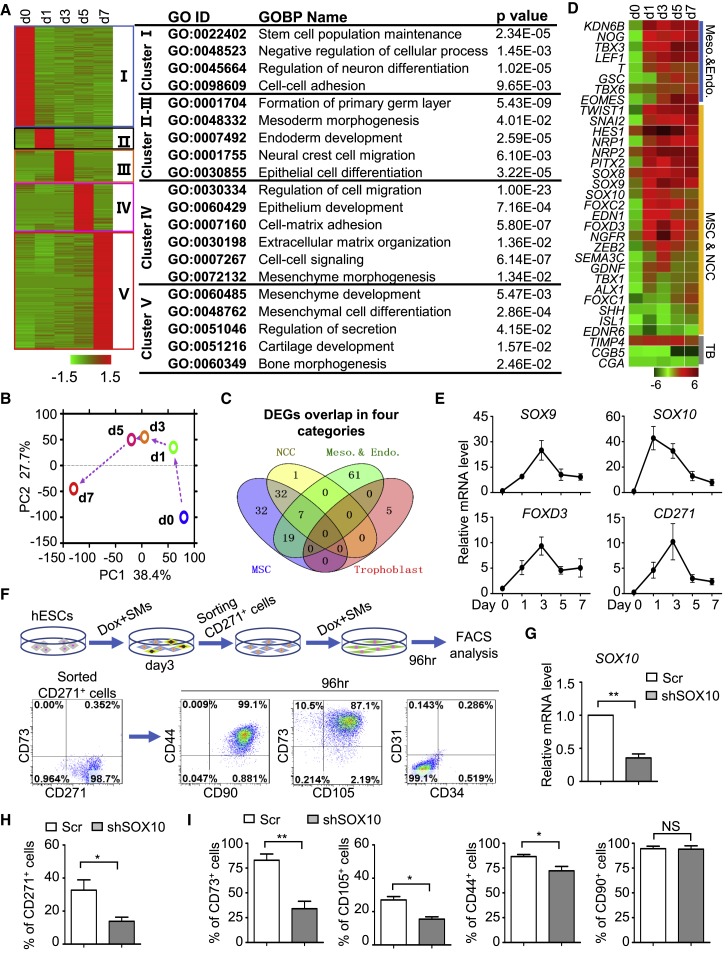
(A) Hierarchical cluster analysis of 2,782 differentially expressed genes (DEGs) (fold change >2, fragments per kilobase of transcript per million mapped reads >0.2) in whole-transcriptome level (left) and GO biological process (GOBP) analysis with p value (right).
(B) PCA of samples of MC-MSC induction from GFP-MSX2 H1 hESCs. PC1, principal component 1; PC2, principal component 2.
(C) Venn diagram shows the overlap among MSC, NCC, mesoderm and endoderm, and trophoblast-associated genes enriched in 14,453 DEGs.
(D) Heatmap illustration shows the expression changes of mesendoderm, MSC and NCC, and TB (trophoblast)-associated genes during MC-MSC induction.
(E) qRT-PCR analysis of the dynamic expression for NCC-associated genes during MSC induction from GFP-MSX2 H1 hESCs with DOX + SM (mean ± SEM, N = 3).
(F) MSC potential analysis of CD271+ NCCs derived from GFP-MSX2 H1 hESCs. CD271+ cells were isolated at day 3 of differentiation and cultured with DOX + SM for 96 hr, followed by FCM analysis.
(G) qRT-PCR analysis showing the depletion of SOX10 by small hairpin RNAs (shRNAs; a mixture of shSOX10-1, shSOX10-2) (mean ± SEM, N = 3). ∗∗p < 0.01.
(H and I) FCM analysis for CD271+ NCCs at day 3 (H) or MSC markers at day 7 (I) in H1 hESCs (Scr, shSOX10) under SMs conditions, respectively (mean ± SEM, N = 3). ∗p < 0.05; ∗∗p < 0.01; NS, not significant.
See also Figure S4.