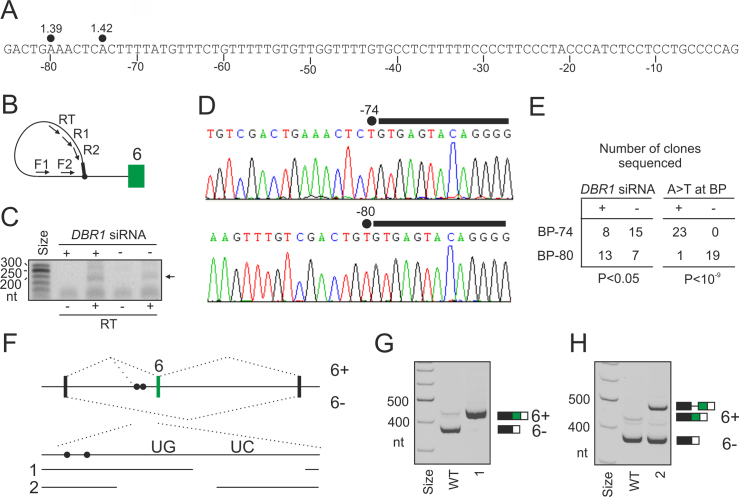
Figure 8.
Functional and structural PPT partitioning. (A) Nucleotide sequence upstream of GANAB exon 6. Predicted BP adenines are denoted by closed circles; numbers indicate their SVM scores (49). B, BP mapping primers (Supplementary Table S1). The 5′ end of intron is denoted by a black rectangle. C, PCR products amplified from DBR1-depleted (+) and control (–) cultures. Samples were reverse-transcribed in the presence (RT+) or absence (RT–) of reverse transcriptase. Products shown in panel D are denoted by an arrow. (D) Representative sequence chromatograms showing A>T mismatches at the lariat junction, which occurs when RT traverses the noncanonical 2′ to 5′ linkage between the 5′ss nucleotide and BP (66,92). (E) Distribution of BPs mapped to positions –74 and –80 in DBR1-depleted cells and controls (left panel) and with/without A>T substitutions at the lariat junction (right panel). P-values were derived from Fisher's exact tests. (F) Deletions of the UC- and UG-rich segments in the PPT of the WT GANAB reporter construct (deletion 1 and 2). Closed circles show two BPs mapped in panels B–D. (G, H) Splicing pattern of the two deletion constructs after transient transfection into HEK293 cells. RNA products are to the right. Cryptic 3′ss activated by deletion 2 is 91 nt upstream of the natural 3′ss of exon 6 and is schematically shown in panel F.