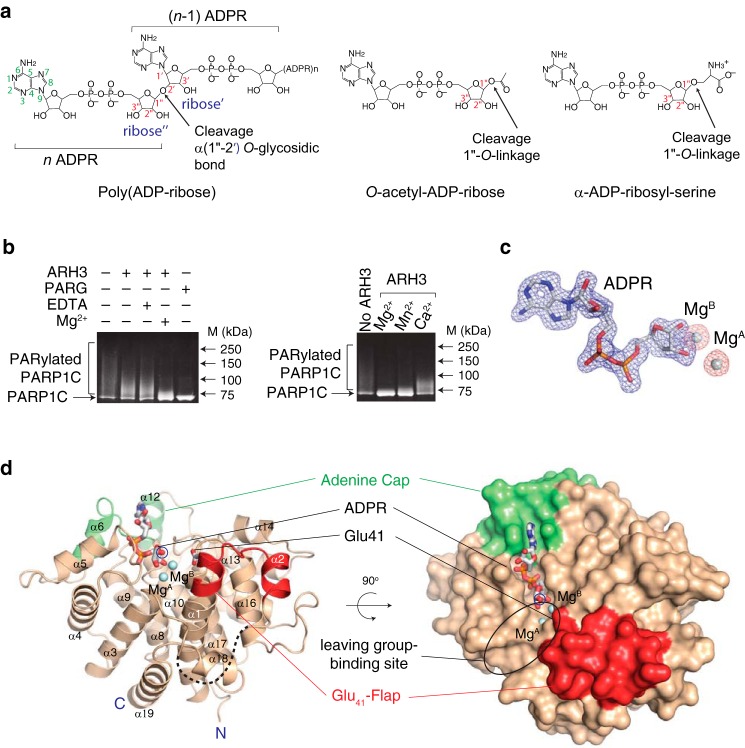
Figure 1.
Structure of full-length human ARH3 bound to ADP-ribose and Mg2+. a, structures of ARH3 substrates: a linear unbranched poly(ADP-ribose) (left), O-acetyl-ADP-ribose (middle), and α-ADP-ribosylserine (right). ARH3 cleaves the 1″-O-linkage in substrates. The exoglycohydrolase activity of ARH3 cleaves the α(1″-2′) O-glycosidic bond between n and n − 1 ADP-ribose, releasing ADP-ribose as a product. b, left, Mg2+ enhances the ADP-ribosyl-acceptor hydrolase activity of ARH3. The ARH3-mediated hydrolysis of PAR on PARylated PARP1C was monitored in the presence and absence of Mg2+ using a gel-based assay. PARG has stronger PAR turnover activity than ARH3 and was used as a positive control. Right, metal preference of ARH3. ARH3 prefers Mg2+ for catalysis followed by Mn2+ and Ca2+. c, difference electron density maps (Fo − Fc) for ADPR and Mg2+ ions contoured at 3.0 σ (blue, ADPR; orange, Mg2+). d, structure of the ARH3–ADPR–Mg2+ complex revealing two unique and flexible structural elements, adenine cap (green) and glutamate flap (Glu41-flap) (red), that undergo conformational changes and strongly contribute to specific substrate recognition. The 1″-OH of ADPR (blue circled), corresponding to the scissile 1″-O-linkage in substrates, is exposed to solvent, consistent with ARH3 specificity for 1″-O-linkage for cleavage. A putative binding site for the leaving group is highlighted with a black ellipse. The N and C termini of ARH3 are indicated by N and C, respectively.