Figure 2.
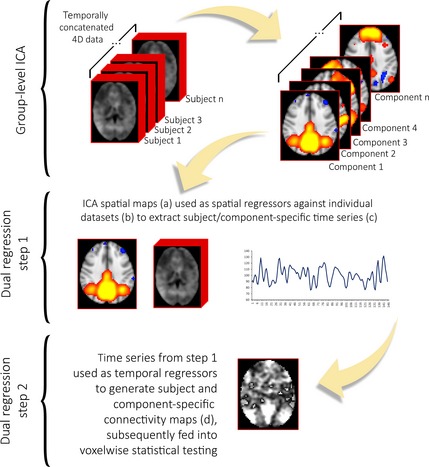
Independent component analysis (ICA) is a data‐driven procedure that identifies coherent spatial signal fluctuation patterns in the dataset, extracting maximally independent components associated with the underlying signal sources—such as ICN and spatially structured artifacts—while avoiding the potential biases in the a priori selection of ROIs 105, 107. The number of components estimated in ICA (i.e., its dimensionality reduction) is a possible source of variability in study results as there is no single best approach for characterizing the complex hierarchy of ICN neurobiology 104. Performing between‐subject ICA analysis is not a straightforward procedure, as it is difficult to establish a direct, one‐to‐one correspondence of ICNs identified with individual‐level ICAs 108. Most current resting‐state fMRI approaches involve performing group‐level ICA on the temporally concatenated datasets of all subjects—allowing the extraction of subject‐specific time courses and group‐common spatial maps 108. This is followed by the reconstruction of individual ICN maps through procedures such as dual regression or direct back‐reconstruction techniques 104, 108, 109, 110. These methods minimize the problem of intersubject ICN correspondence and takes advantage of the higher signal‐to‐noise ratio offered by analyzing several subjects conjointly 104.
