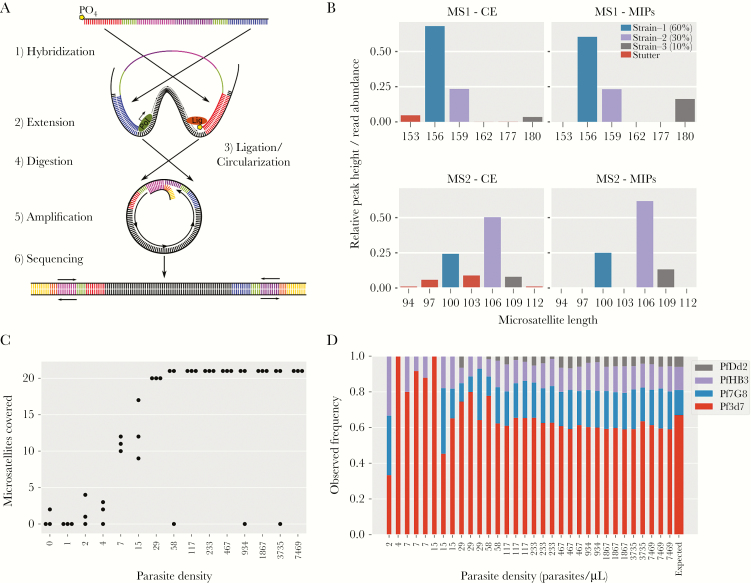
Figure 1.
Molecular inversion probe (MIP) assay and performance on laboratory control mixtures. A, The MIP capture is illustrated showing the key steps of MIP arm hybridization, polymerase extension, and gap ligation to form a single-stranded circle. Exonuclease digestion removes linear template DNA, thereby relatively enriching for the circular captures, which are then amplified using universal primers along with a sample barcode. Important components are color coded: extension arm (blue), ligation arm (red) molecular identifiers (green), and backbone (pink+purple). B, An example of microsatellite (MS) stutter seen in standard capillary electrophoresis versus MIPs where stutter is detected and removed based on inconsistency within unique molecular identifiers. C, The coverage of the 21 assessed MSs, demonstrating that apart from a few failed reactions the vast majority of MSs are detected in every sample until dilutions of 29 parasites/uL. D, Frequency estimates of the 4-strain mixture compared with expected frequency (last bar on right, drawn wider for emphasis) based on relative amounts of DNA from each strain. Abbreviations: CE, capillary electrophoresis; MIP, molecular inversion probe; MS, microsatellite.