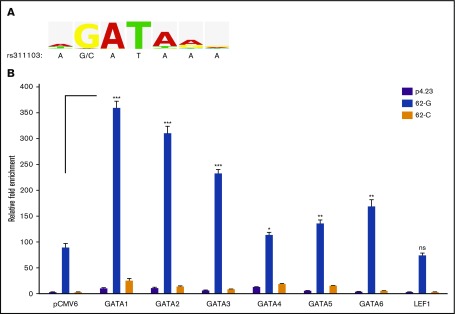
Figure 5.
Effects of the erythroid GATA factors on the transcriptional activity of rs311103[G] genomic region. (A) Consensus sequence for the GATA transcription factor family. The sequence logo, derived from the V$GATA_Q6 matrix report in TRANSFAC, graphically displays the nucleotide frequencies in the binding motif of the GATA transcription factor family. The genomic sequence spanning SNP rs311103 is shown below. The V$GATA_Q6 matrix was constructed based on 105 sequence entries for the genomic binding sites of various GATA1-GATA6 factors from human, mouse, rat, and chicken, and the constructed binding motif of the matrix consisted of a consensus sequence, WGATAR, for the GATA family. The G nucleotide at the second position of this consensus occurred at a high frequency of 98% (103 sequence entries), whereas none of the 105 sequence entries had the C nucleotide at that position. (B) Effects of the GATA family, GATA1-GATA6, and LEF1 transcription factors on the transcriptional activities of 62-G and 62-C reporter constructs. The expression vectors for GATA1-GATA6 and LEF1 and the empty pCMV6 vector were cotransfected with the 62-G or 62-C reporter constructs or the empty pGL4.23 vector (p4.23) into the host K-562 cells. Relative luminescence measurements obtained from each experiment were normalized against β-galactosidase activity. The results are presented as the fold change in the mean of luminescence units obtained from triplicate experiments relative to that obtained from the cells transfected with empty pCMV6 and pGL4.23 vectors; the error bars indicate the standard deviations. Statistical analyses between the 62-G reporter result obtained from the pCMV6 control group and each of the 62-G reporter results obtained from the GATA1-GATA6 and LEF1 expression groups were performed using the unpaired t test with the Welch correction. ***P <.001, **P <.01, *P < .05. ns, nonsignificant.