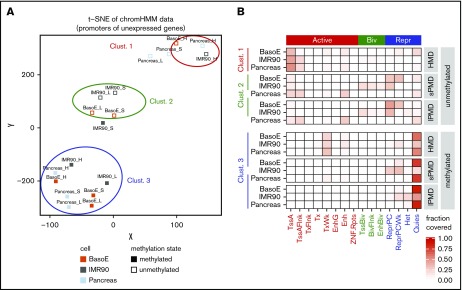
Figure 4.
Transcriptionally inactive promoters are regulated by different mechanisms in HMDs and PMDs. (A) Graph illustrating t-SNE analysis of the chromatin status as determined by ChromHMM of silent IMR90, pancreas, and BasoE promoters, separated by their methylation status and their location in PMDs or HMDs. The t-SNE was used with default parameters and a perplexity of 4. Inactive promoters from BasoEs, IMR90, and pancreas were divided into 6 categories: methylated (open squares) and unmethylated (closed squares) located in either HMDs (H), s-PMDs (S), or l-PMDs (L). Multiple runs of t-SNE for all inactive promoters, as well as analyses using only the inactive CpG-rich promoters resulted in very similar clusters (not shown). Inactive promoters fell into 3 clusters: cluster 1 contained unmethylated promoters located in HMDs, cluster 2 contained unmethylated promoters located in short and long PMDs, and cluster 3 contained all methylated promoters (except for the IMR90 s-PMDs). (B) Heat map illustrating the chromatin status (based on ChromHMM) of promoters in clusters 1, 2, and 3. The fractions of promoter overlapping the indicated ChromHMM state are shown. Promoters in cluster 1 generally exhibited active chromatin marks, whereas those in cluster 2 exhibited repressive or bivalent marks, demonstrating that the mechanisms of silencing of unmethylated promoters in HMDs and PMDs are different. Promoters in cluster 3 were almost all quiescent regardless of their location in HMDs or PMDs.