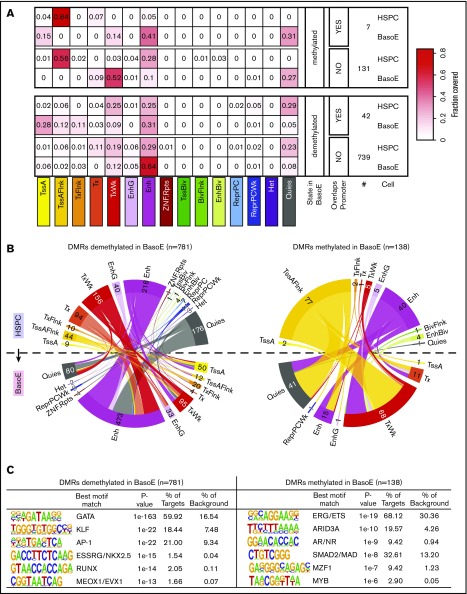
Figure 6.
DNA methylation and chromatin changes during erythroid differentiation in DMRs. (A) Heat maps illustrating the changes in chromatin state of short DMRs within HMDs between HSPCs and BasoEs. Top and bottom panels show average fractional chromatin composition by sequence length of DMRs that are more or less methylated in BasoEs than in HSPCs, respectively. They are additionally broken down into DMRs that overlap promoters and those that do not. The numbers indicate the number of DMRs in each category. The 15 ChromHMM states are indicated below the heat map. Most notable changes include disappearance of the flanking active TSS (TssAFlnk) marks and increase of enhancer marks between HSPCs and BasoEs. (B) Circos plots illustrating the changes in chromatin state of DMRs that lose (left) or gain (right) methylation during differentiation from HSPCs (top half of the circle) to BasoEs (bottom half). The bands connecting the half-circles indicate the redistribution of DMRs from HSPCs into the indicated ChromHMM categories in BasoEs. The numbers indicate the number of DMRs in each category. Again, the most notable changes include the increase in the enhancer marks and reduction of quiescence marks upon demethylation of DMRs upon differentiation (left), and, conversely, the increase in quiescence marks and weak transcription and the decrease of chromatin marks associated with enhancers and transcriptional flanks upon gain of methylation in DMRs between HSPCs and BasoEs (left). (C) Top enriched sequence motifs in DMRs identified by Homer motif analysis. Best motif matches to the identified motifs shown as positional weight matrices (PWMs) are indicated, and enrichment parameters for each motif are shown. DMRs demethylated in BasoEs were enriched in erythropoietic transcription factors (right), whereas DMRs that gained methylation in BasoEs were enriched in genes associated with stem cells and lymphoid cells, consistent with lineage restriction-associated changes in enhancer methylation.