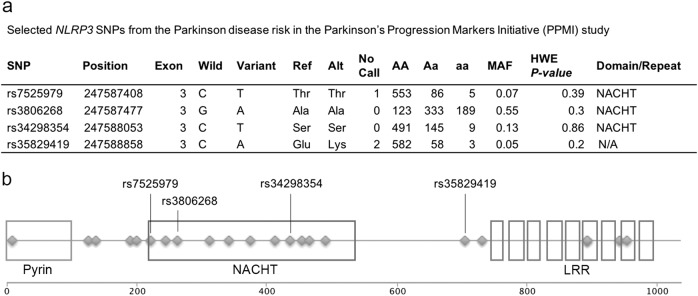
Fig. 2.
Genetic variation of NLRP3. a Whole-exome sequences of individuals in the PPMI database (n = 644: 402 PD patients, 60 SWEDD, and 182 controls) were assessed. A total of 22 SNPs in NLRP3 were identified. Four SNPs with an MAF ≥ 5% in this cohort were selected for further analysis rs7525979, rs3806268, rs34298354, and rs35829419. Table header abbreviations: SNP single nucleotide polymorphism, Position nucleotide position within the genome, Broad Institute, Exon location of SNP, Wild WT nucleotide, Variant variant nucleotide, Ref amino acid coded for by WT nucleotide, Alt amino acid coded for by inclusion of variant nucleotide, MAF minor allele frequency, HWE P value Hardy-Weinberg Equilibrium analysis P value, Domain/Repeat location of SNP within protein. b Schematic representation of the NLRP3 gene structure. NLRP3, located on the long arm of chromosome 1, codes for a multi-functional NOD-like receptor (NLR) protein containing a highly conserved NACHT domain along with an n-terminal pyrin domain and a c-terminal leucine-rich repeat domain. The protein is comprised of 1036 amino acids and has a molecular mass of 118 kDa. Three of the four NLRP3 SNPs identified in this study that have a MAF ≥ 5% are located in the NACHT domain. All SNPS with a MAF ≥ 5% are indicated with respective Reference SNP numbers. All 22 NLRP3 SNPs identified in the PPMI database are indicated with a diamond along the protein diagram