Abstract
Background
Age-related gait speed decline is accelerated in men with human immunodeficiency virus (HIV). Mitochondrial genetic variation is associated with frailty and mortality in the general population and may provide insight into mechanisms of functional decline in people aging with HIV.
Methods
Gait speed was assessed semiannually in the Multicenter AIDS Cohort Study. Mitochondrial DNA (mtDNA) haplogroups were extracted from genome-wide genotyping data, classifying men aged ≥50 years into 5 groups: mtDNA haplogroup H, J, T, Uk, and other. Differences in gait speed by haplogroups were assessed as rate of gait speed decline per year, probability of slow gait speed (<1.0 m/s), and hazard of slow gait using multivariable linear mixed-effects models, mixed-effects logistic regression models, and the Andersen-Gill model, controlling for hepatitis C virus infection, previous AIDS diagnosis, thymidine analogues exposure, education, body composition, smoking, and peripheral neuropathy. Age was further controlled for in the mixed-effects logistic regression models.
Results
A total of 455 HIV-positive white men aged ≥50 years contributed 3283 person-years of follow-up. Among them, 70% had achieved HIV viral suppression. In fully adjusted models, individuals with haplogroup J had more rapid decline in gait speed (adjusted slopes, 0.018 m/s/year vs 0.011 m/s/year, pinteraction = 0.012) and increased risk of developing slow gait (adjusted odds ratio, 2.97; 95% confidence interval, 1.24–7.08) compared to those with other haplogroups.
Conclusions
Among older, HIV-infected men, mtDNA haplogroup J was an independent risk factor for more rapid age-related gait speed decline.
Keywords: mitochondrial genetic, HIV, aging, gait speed
Mitochondrial DNA haplogroup and longitudinal gait speed decline after age 50 years were studied among white men infected with human immunodeficiency virus. Haplogroup J was an independent risk factor for more rapid age-related gait speed decline.
Due to effective antiretroviral therapy (ART), people living with human immunodeficiency virus (PLWHIV) have substantially longer life expectancies than in the pre-ART era [1, 2], yet have higher rates of chronic comorbidities and greater risk of premature death than demographically similar adults without HIV [3, 4]. Accelerated functional decline and higher prevalence of frailty among PLWHIV have been observed compared to HIV-uninfected controls [5–8]. Gait speed is an independent measure of physical function and a component of the assessment of frailty and has been shown to predict functional decline, hospitalization, disability, and deaths among older adults [6, 9]. An accelerated rate of decline in gait speed has been noted in older PLWHIV, which could be attributed to multiple factors, including characteristics shared with the general population (eg, reduced energy production [10], compromised energy consumption [10, 11], changes in body composition [12]), as well as unique challenges from HIV chronic infection [13, 14].
Mitochondria, which actively engage in cell energy production [15], are likely an important contributor to functional aging in PLWHIV. Mitochondrial DNA (mtDNA) is associated with energy production, apoptosis, and inflammation pathways in cells [16] and has been linked to age-related physical function decline [15, 17], frailty, and mortality [18]. Conversely, mitochondrial function among PLWHIV could be damaged by HIV chronic infection [14] and long-term use of ART [13], which trigger proinflammatory responses that could lead to impaired physical function. It is possible that mtDNA genetic variations interact with multiple mechanisms and confer additional susceptibility to certain individuals.
Mitochondrial DNA haplogroups represent major branch points on the mitochondrial phylogenetic tree and have been broadly used in studying human evolution and discovering genes involved in complex disease development [19]. Among the HIV-infected (HIV-positive) population, mtDNA haplogroups have been linked to progression to AIDS and mortality [20], as well as ART-associated peripheral neuropathy [21]; however, their association with physical function in PLWHIV has not been investigated. Our aim in this study was to evaluate the association of common European mtDNA haplogroups with gait speed in PLWHIV to better understand the role of mitochondrial genetic background in aging-related declines in physical function.
METHODS
Study Population
We used data from the Multicenter AIDS Cohort Study (MACS), a long-standing prospective community-based cohort that recruited men with and without HIV who have sex with men at 4 study sites (Baltimore/Washington, DC, Chicago, Pittsburgh, and Los Angeles) starting in 1984. Institutional review boards at each study location approved the study protocol, and each participant provided informed written consent. Descriptions of the study design, enrollment, and data collection can be found elsewhere [6, 22]. Briefly, participants were enrolled during 3 enrollment periods from 1984–1985, 1987–19891, and 2001–2003 and attend routine semiannual visits for standardized interviews, laboratory tests, and physical examinations. For purposes of this analysis, participants were restricted to non-Hispanic white males, because the greatest proportion of MACS participants available for analysis were of non-Hispanic white race/ethnicity. We only included participants who had 2 or more visits with physical function measurement and had genetic data available. No mtDNA haplogroup information was available for HIV-negative men at the time of analysis, thus we only included HIV-positive men in the current study. Furthermore, we restricted this analysis to men aged ≥50 years, because a previous MACS study demonstrated an accelerated rate of gait speed decline after age 50 among HIV-positive participants [6].
Gait Speed
Gait speed was assessed semiannually in meters per second over a 4-meter course using standard clinical procedures [23]. Participants were asked to walk at their “normal, comfortable pace.” Timing was initiated with a command of “Go” and stopped after the first foot-fall over the finish line. Two measurements were conducted, with the faster measurement [24] used for analysis. A measurement of <1 m/s was defined as clinically slow gait speed [23, 25]. Gait speeds >1.7 m/s were recorded as missing (52 visits out of 4031) in the analysis due to potential measurement error, as 1.7 m/s may be considered a transitional speed between walking and running [26, 27].
HIV Serostatus, HIV Disease Progression, and ART Use
All men in this analysis were HIV positive, tested by enzyme-linked immunosorbent assay and confirmed by Western blot. HIV disease progression was tracked through history of AIDS and log10 plasma HIV viremia copy-year. HIV viral load was measured using the Roche ultrasensitive assay (limit of detection = 50 copies/mL; Roche Diagnostics, Nutley, New Jersey). HIV viremia copy-year is a time-varying measurement of cumulative HIV burden since seroconversion [28, 29] and is used to estimate the area under the patient’s longitudinal viral load curve from seroconversion to each visit. In the current study, we calculated viremia copy-year for those with and without HIV at baseline using their viral load measured since 2003 or since seroconversion, respectively. Nucleoside reverse transcriptase inhibitors (NRTIs), especially thymidine analogues NRTI, has known mitochondrial toxicity [30]. Therefore, we controlled for time-varying exposure to thymidine analogues (stavudine [d4T] and Zidovudine [AZT]) in the final models.
mtDNA Haplogroup Determination
mtDNA genotyping data were extracted from multiple existing genome-wide genotyping panels (Illumina Human Hap 550, Illumina 1MDuo, Illumina 1M, and TaqMan) in MACS. Comprehensive quality control was completed using PLINK software. All heterozygous calls were set to be missing, and single-nucleotide polymorphism (SNP) strands were flipped to match the mitochondrial reference. mtDNA haplogroups were identified using HaploGrep [19], an application that identifies the most probable mtDNA haplogroups based on the phylogenetic tree of global mtDNA variation (Phylotree: http://www.phylotree.org) [31]. The vast majority of study participants fit into 1 of the 4 common European haplogroups: H, J, T, and Uk; the remaining were combined into the “other” haplogroup.
Other Covariates
Data on sociodemographic status (age, race, and whether or not they have college education) and cigarette smoking (time-varying never vs ever smoker, time-varying never vs previous vs current smoker, or pack-years of smoking) were collected through interviews during enrollment and at each study visit. Hepatitis C virus (HCV) infection was identified as anti-HCV seropositive. Peripheral neuropathy was defined as ever reporting pain, burning, numbness, a pins-and-needles sensation in the feet or legs, or inability to detect vibrations in either foot [6].
Statistical Analyses
All statistical analyses were conducted using Stata software, version 14 (StataCorp, College Station, Texas). Baseline characteristics of participants were stratified by 5 groups (haplogroup H, J, T, Uk, and other), and differences were evaluated using Fisher exact test for categorical variables and Kruskal-Wallis equality-of-populations rank test for continuous variables.
During the exploratory data analysis, scatter plots, linear regression fitted lines (Supplementary Figure 1), and locally weighted regression smoothers were used to compare gait speed decline for each haplogroup against all other haplogroups. Using longitudinal data, changes in gait speed were evaluated among men from different haplogroups. Specifically, we conducted the following analyses by haplogroups: (i) The rate of gait speed decline (change in meters per second per year) was determined using random effects mixed linear models with random slopes and intercepts after controlling for the following variables: HIV viremia copy-year, ever diagnosed with AIDS, HCV serostatus, weight (kilograms), height (meters), ever used any thymidine analogues, peripheral neuropathy (yes/no), college education (yes/no), and time-varying smoking status (never vs former vs current). Since we did not observe baseline difference in gait speed by haplogroups, random effects mixed linear models were constructed with the assumption that there was no baseline difference. (ii) The odds of slow gait speed (<1.0 m/s) were identified using mixed effects logistic regression models after controlling for the same covariates. The time unit for longitudinal analysis (i and ii) was 1 year, starting at age 50. Because participants could have more than 1 slow gait speed visit, we estimated (iii) hazard ratios (HRs) of multiple slow gait visits using the Andersen-Gill model [32]. Covariates in adjusted models were selected based on their statistical significance (P < .05) and change of magnitude in the association of interest.
RESULTS
Baseline Characteristics
The study sample included 455 HIV-positive, self-reported non-Hispanic white men aged ≥50 years who had 2 or more study visits between 1 October 2007 and 30 September 2016. They contributed a total of 3283 person-years of follow-up to the analysis (median follow-up, 8 person-years; interquartile range, 4–11). Table 1 lists characteristics of participants at baseline (first visit after 1 October 2007 when participants were aged >50) by mtDNA haplogroups. Median age was 50 years, the majority (58.7%) had college education or higher, mean CD4+ cell count was >500 cells/µL, and most (69.5%) had a suppressed HIV-1 RNA.
Table 1.
Baseline Characteristics by Mitochondrial DNA Haplogroups
| Mitochondrial DNA Haplogroups | |||||||
|---|---|---|---|---|---|---|---|
| Characteristic | Overall | H | J | T | Uk | Others | P Valuea |
| Overall, N (%) | 455 | 188 (41.3) | 44 (9.7) | 53 (11.7) | 108 (23.7) | 62 (13.6) | … |
| Age, median (IQR), year | 50 (50–54) | 50 (50–53) | 51 (50–54.5) | 50 (50–54) | 50.5 (50–54) | 52 (50–54) | .11 |
| Have college degree, N (%) | 267 (58.7) | 100 (53.2) | 26 (59.1) | 34 (64.2) | 69 (63.9) | 38 (61.3) | .36 |
| Ever smoke, N (%) | 307 (70.7) | 119 (68.0) | 34 (79.1) | 42 (79.3) | 72 (68.6) | 40 (69.0) | .38 |
| Hepatitis C virus infection, N (%) | 75 (16.5) | 29 (15.4) | 6 (13.6) | 16 (30.2) | 16 (14.8) | 8 (12.9) | .11 |
| CD4 cell count at baseline, median (IQR), cells/µL | 523 (354–727) | 499 (358–689) | 550 (323–795) | 597 (371–819) | 533 (371–743) | 510 (325–689) | .74 |
| Human immunodeficiency virus virally suppressed (<200 copies/mL), N (%) | 316 (69.5) | 129 (68.6) | 28 (63.6) | 39 (73.6) | 72 (66.7) | 48 (77.4) | .49 |
| Peripheral neuropathy, N (%) | 170 (37.4) | 70 (37.2) | 16 (36.4) | 19 (35.8) | 44 (40.7) | 16 (35.6) | .96 |
| Weight, median (IQR), kg | 78.6 (70.8–86.2) | 78.9 (72–85.5) | 79.4 (69.1–91) | 79.5 (70.8–86.6) | 76.3 (69.5–85.3) | 79.8 (71.7–85.3) | .69 |
| Height, median (IQR), m | 1.78 (1.73–1.83) | 1.78 (1.73–1.83) | 1.80 (1.75–1.80) | 1.78 (1.73–1.80) | 1.78 (1.73–1.83) | 1.80 (1.75–1.85) | .17 |
| Ever used thymidine analogues, N (%) | 51 (11.5) | 21 (11.7) | 3 (7.0) | 7 (13.2) | 10 (9.4) | 10 (17.0) | .54 |
| Gait speed, mean (standard deviation), m/s | 1.15 (0.19) | 1.13 (0.17) | 1.07 (0.21) | 1.20 (0.21) | 1.20 (0.23) | 1.19 (0.19) | .29 |
Abbreviation: IQR, interquartile range.
aCategorical variables tested by Fisher exact test; continuous variables tested by Kruskal-Wallis equality-of-populations rank test.
As presented in Table 1, H was the most prevalent haplogroup (41%), followed by Uk, T, and J. Baseline demographic and behavioral characteristics were similar by haplogroup. Mean gait speed at baseline visit was 1.15 m/s overall and was slightly slower for participants with haplogroup J (not statistically significant, P = 0.29).
Gait Speed Decline and mtDNA Haplogroups
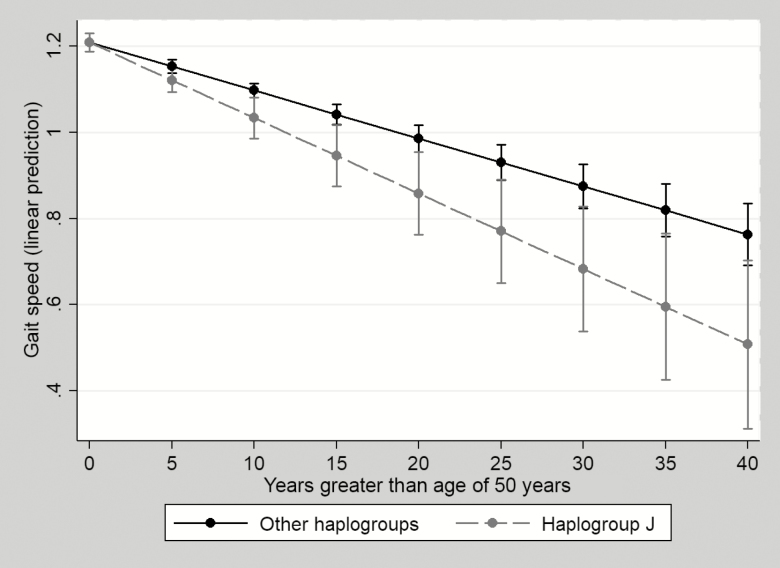
In 4 random effects mixed linear models (Table 2), all haplogroups had significant declines in gait speed over time. However, the interaction between haplogroup and time was only significant in the model with haplogroup J, suggesting the rate of gait speed decline over time was only significantly different when comparing haplogroup J vs non-J (P value for interaction between time and haplogroup, 0.012). As reflected in Table 2 and Figure 1, gait speed declined at a rate of 0.018 m/s/year among haplogroup J vs 0.011 m/s/year in non-J, after controlling for covariates. Consistent with our previous study performed in MACS [6], HIV viral suppression did not have a significant effect on gait speed. However, having ever been diagnosed with AIDS and higher viremia copy-year were associated with more rapid decline of gait speed (Supplementary Table 1). To prevent bias estimation from dichotomizing exposure (haplogroup) in mixed effect linear regression models, we also constructed a mixed effect linear regression model that fit all haplogroups as a categorical variable and assessed the trajectory of gait speed decline in each haplogroup compared to haplogroup H (Supplementary Table 2). All estimations were similar to the results presented in Table 2.
Table 2.
Unadjusted and Adjusted Longitudinal Association between Age and Gait Speed by Mitochondrial DNA Haplogroups
| Models | Independent Variables | Crude Estimation | Adjusted Estimationa | ||||
|---|---|---|---|---|---|---|---|
| Slope (m/s/year) | SE | P Value | Slope (m/s/year) | SE | P Value | ||
| Haplogroup J vs others | J*age | –0.006 | 0.003 | .026 | –0.006 | 0.003 | .012 |
| Gait speed (non-J) | –0.012 | 0.001 | <.001 | –0.011 | 0.001 | <.001 | |
| Gait speed (group J) | –0.017 | 0.003 | <.001 | –0.018 | 0.003 | <.001 | |
| Haplogroup H vs others | H*age | 0.001 | 0.002 | .48 | 0.002 | 0.002 | .20 |
| Gait speed (non-H) | –0.013 | 0.001 | <.001 | –0.012 | 0.001 | <.001 | |
| Gait speed (group H) | –0.011 | 0.001 | <.001 | –0.010 | 0.001 | <.001 | |
| Haplogroup T vs others | T*age | 0.00001 | 0.002 | .996 | –0.00007 | 0.002 | .98 |
| Gait speed (non-T) | –0.012 | 0.001 | <.001 | –0.012 | 0.001 | <.001 | |
| Gait speed (group T) | –0.012 | 0.002 | <.001 | –0.012 | 0.002 | <.001 | |
| Haplogroup Uk vs others | Uk*age | –0.0005 | 0.002 | .78 | 0.0001 | 0.002 | .94 |
| Gait speed (non-Uk) | –0.012 | 0.001 | <.001 | –0.012 | 0.001 | <.001 | |
| Gait speed (group Uk) | –0.013 | 0.001 | <.001 | –0.012 | 0.002 | <.001 | |
Gait speed modeled as a continuous variable in meters per second. Boldface indicates statistical significance for the interaction between mitochondrial DNA haplogroup and age.
Abbreviation: SE, standard error.
aModels adjusted for hepatitis C virus infection, AIDS diagnosis, log10 viremia copy-year, college education, cigarette smoking, peripheral neuropathy, weight, height, and time-varying exposure to thymidine analogues (ever vs never). Cigarette smoking was controlled in models by time-varying smoking status (yes vs no), time-varying smoking status (never vs former vs current), or pack-year smoke. All methods returned similar changes of the magnitude of association between haplogroup and gait speed. Time-varying smoking status (never vs former vs current) was selected and used in the final model.
Figure 1.
Adjusted predictions of gait speed decline by haplogroup J with 95% confidence intervals. Statistics presented in Table 2 and Supplementary Table 1. Model adjusted for hepatitis C virus infection, AIDS diagnosis, log10 viremia copy-year, college education, cigarette smoking, peripheral neuropathy, weight, height, and time-varying exposure to thymidine analogues (ever vs never).
Probability of Slow Gait Speed by Haplogroup J
Mixed effects logistic regression models were used to examine differences in the probability of developing clinically slow gait speed between haplogroup J and non-J. Compared to non-J haplogroups, having haplogroup J was associated with a 2.97-fold (95% confidence interval [CI], 1.24–7.08) higher risk of developing slow gait speed, after controlling for covariates (Table 3). The HR of slow gait speed among group J vs non-J from the Andersen-Gill model confirmed these findings (adjusted HR, 1.60; 95% CI, 1.04–2.47).
Table 3.
Probability of Slow Gait after Age of 50 Years by Independent Variables
| Independent Variables | Crude Estimation | Adjusted Estimation | ||||
|---|---|---|---|---|---|---|
| OR | 95% CI | P Value | OR | 95% CI | P Value | |
| Haplogroup J | 3.35 | 2.37–8.86 | .015 | 2.97 | 1.24–7.08 | .01 |
| Age | 1.15 | 1.11–1.21 | <.001 | 1.14 | 1.10–1.19 | <.001 |
| Hepatitis C virus infection | … | … | … | 1.29 | 0.56–2.97 | .55 |
| Ever diagnosed with AIDS (yes/no) | … | … | … | 1.89 | 1.04–3.46 | .04 |
| Log10 viremia copy-year, per unit increase | … | … | … | 1.25 | 1.03–1.52 | .02 |
| College education (yes/no) | … | … | … | 0.34 | 0.20–0.60 | <.001 |
| Ever smoke | ||||||
| Never | … | … | … | Ref. | … | … |
| Former | … | … | … | 1.24 | 0.67–2.28 | .49 |
| Current | … | … | … | 1.30 | 0.64–2.63 | .46 |
| Weight, kg | … | … | … | 1.0 | 0.99–1.01 | .75 |
| Height, m | … | … | … | 0.04 | 0.0005–3.66 | .17 |
| Exposure to thymidine analogues (ever vs never) | … | … | … | 0.44 | 0.17–1.15 | .10 |
| Peripheral neuropathy (yes/no) | … | … | … | 1.33 | 1.01–1.76 | .04 |
Abbreviations: CI, confidence interval; OR, odds ratio.
DISCUSSION
To our knowledge, this is the first study to evaluate the contribution of mitochondrial genetics to physical function in PLWHIV. We found that although there was no difference in gait speed at age 50, gait speed declined significantly faster among participants with haplogroup J than among participants with other haplogroups. In other studies, a minimum clinically meaningful difference in gait speed has generally been taken to be between 0.05 and 0.10 m/s [33]. Studenski et al conducted pooled analysis of 9 large cohorts and suggested that the hazard of deaths for older adults decreased by 12% (95% CI, 0.87–0.90) with each increase of 0.1 m/s in gait speed [9]. According to our results, men with haplogroup J, declining 0.006 m/s faster per year than men with other haplogroups, would reach a difference of 0.05–0.1 m/s after 8 to 16 years (ie, as early as age 58). Also, participants in haplogroup J had a 2.97-fold increased risk of having a study visit with slow gait speed (<1 m/s). Both faster decline in gait speed and higher probabilities of slow gait among haplogroup J were independent from participants’ HIV disease state and exposure to thymidine analogues. These findings support the hypothesis that inherited mtDNA variation is associated with physical function decline among PLWHIV.
Previous studies linking mtDNA haplogroups and health outcomes have also noted issues common to haplogroup J. Haplogroup J is known to be associated with increased penetrance of Leber hereditary optic neuropathy [34, 35], possibly due to mtDNA defects. Gómez-Durán et al observed that compared to haplogroup H, cytoplasmic hybrids from haplogroup J contain less mtDNA and RNA, synthesized a smaller amount of mtDNA-encoded polypeptides, and also displayed lower oxygen consumption, lower mitochondrial inner membrane potential, and lower total ATP levels [36], suggesting less efficient mitochondrial function. Lower energy production and reduced energy consumption are associated with faster physical function decline in the general population [10, 11]. In contrast, Suissa et al observed that individuals with haplogroup J had a 2-fold increase in mtDNA copy number compared to those with haplogroup H [37]. It is possible that such an increase might be due to compensatory response to mitochondrial function damage. Further studies should investigate associations between mtDNA genetics and mitochondrial heteroplasmy to verify these findings.
Within the HIV-positive population, haplogroups J and U5 have been observed to be more common among those who display accelerated progression to AIDS and death [20]. The study suggested that both haplogroups (J and U5) contained uncoupling SNPs and were associated with lower ATP production and reactive oxygen species generation compared to other haplogroups (eg, H3, H4, H5, and H6) that were more tightly coupled [20], suggesting mtDNA haplogroups could be linked to mitochondrial function variation and play a role in AIDS progression. Our findings suggest that even among men who have achieved HIV viral suppression, such variants as found among haplogroup J could be associated with physical function decline and predict worse functional outcome in PLWHIV.
Studies linking mtDNA variation and physical function decline in the general population have yielded inconsistent findings. One study examined mtDNA variation in frail and nonfrail older adults and suggested that the mt204 C allele, a site that could affect the efficiency of mtDNA replication [38], was associated with a 2-fold increased odds of frailty and decreased grip strength [18]. In contrast, another study found no associations between any tested mtDNA haplogroup and frailty in a very old adult population [39] but did find that haplogroup K (a subgroup of Uk) was associated with a lower hazard of death (HR, 0.52; 95% CI, 0.30–0.91). Although this study was conducted in a relatively large sample with validated measurements, frailty phenotypes were measured at a baseline age of 85+, which may exceed the age at which frailty manifests [40, 41]. None of these studies prospectively evaluated associations between mtDNA genome and the trajectory of physical function decline.
The current study has several limitations. First, no women were studied, and the haplogroups studied were European ancestry specific. These factors limit the generalizability of our findings. Second, our sample size was relatively small, and the results could be biased by unmeasured confounders, leading to false-positive findings. Also, we could not study subhaplogroups within J, which might have their own specific associations with the outcomes of interest. Further studies are therefore needed to validate our results. Third, very few study visits were captured among individuals with J haplogroup after age 67 (17 observations from 3 unique individuals). Thus, our findings may not be representative of those older than 67. Fourth, we were only able to capture viral load since seroconversion among those who were not seroprevalent at study baseline (35.6%) in the viremia copy-year measurement. Thus, we likely underestimated the viremia copy-year for those infected with HIV before they participated in the study. Last, due to the demands of physically attending study visits, men who were lost to follow-up might be sicker, thus we could have understated the rate of decline in gait speed with age among all haplogroups.
This study has several strengths. We were able to evaluate the rate and trajectory of gait speed based on validated repeated measurements controlling for HIV disease state and thymidine analogues exposure and to quantify differential decline of gait speed in PLWHIV with 4 major European mtDNA haplogroups. Although focused on an HIV-positive population, this study may also improve understanding of the impact of inherited mtDNA on functional aging in the general population. Last, the MACS cohort recruited behaviorally and sociodemographically similar individuals, which minimizes confounding and makes it ideal for the study of genetics.
CONCLUSIONS
Individuals with mitochondrial haplogroup J had more rapid gait speed decline compared to persons with other common European mitochondrial haplogroups. These findings suggest a potential pathway through mtDNA to understand the pathophysiology of accelerated function decline among PLWHIV and may be used to predict risk of function decline in this group if validated prospectively. Investigation into relationships between mitochondrial genetic variation and changes in physical function could bring insights toward development of interventions including personalized long-term disease management strategies that promote healthy aging among PLWHIV.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We acknowledge support from the multicenter AIDS cohort data coordinating center. We also acknowledge support for statistical consultation from the National Center for Research Resources and the National Center for Advancing Translational Sciences (NCATS) of the National Institutes of Health (NIH) (grant 1UL1TR001079).
Disclaimer. The contents of this publication are solely the responsibility of the authors and do not represent the official views of the NIH.
Financial support. Data were collected by the Multicenter AIDS Cohort Study (MACS) with centers at Baltimore (U01-AI35042), the Johns Hopkins University Bloomberg School of Public Health: Joseph B. Margolick (principal investigator [PI]), Jay Bream, Todd Brown, Adrian Dobs, Michelle Estrella, W. David Hardy, Lisette Johnson-Hill, Sean Leng, Anne Monroe, Cynthia Munro, Michael W. Plankey, Wendy Post, Ned Sacktor, Jennifer Schrack, Chloe Thio; Chicago (U01-AI35039), Feinberg School of Medicine, Northwestern University, and Cook County Bureau of Health Services: Steven M. Wolinsky (PI), Sheila Badri, Dana Gabuzda, Frank J. Palella, Jr., Sudhir Penugonda, John P. Phair, Susheel Reddy, Matthew Stephens, Linda Teplin; Los Angeles (U01-AI35040), University of California–Los Angeles Schools of Public Health and Medicine: Roger Detels (PI), Otoniel Martínez-Maza (PI), Peter Anton, Robert Bolan, Elizabeth Breen, Anthony Butch, Shehnaz Hussain, Beth Jamieson, John Oishi, Harry Vinters, Dorothy Wiley, Mallory Witt, Otto Yang, Stephen Young, Zuo Feng Zhang; Pittsburgh (U01-AI35041), University of Pittsburgh, Graduate School of Public Health: Charles R. Rinaldo (PI), James T. Becker, Phalguni Gupta, Kenneth Ho, Lawrence A. Kingsley, Susan Koletar, Jeremy J. Martinson, John W. Mellors, Anthony J. Silvestre, Ronald D. Stall; Data Coordinating Center (UM1-AI35043), the Johns Hopkins University Bloomberg School of Public Health: Lisa P. Jacobson (PI), Gypsyamber D’Souza (PI), Alison Abraham, Keri Althoff, Michael Collaco, Priya Duggal, Sabina Haberlen, Eithne Keelaghan, Heather McKay, Alvaro Muñoz, Derek Ng, Anne Rostich, Eric C. Seaberg, Sol Su, Pamela Surkan, Nicholas Wada; Institute of Allergy and Infectious Diseases: Robin E. Huebner; National Cancer Institute: Geraldina Dominguez. The MACS is funded primarily by the National Institute of Allergy and Infectious Diseases (NIAID), with additional cofunding from the National Cancer Institute, the National Institute on Drug Abuse, and the National Institute of Mental Health. Targeted supplemental funding for specific projects was also provided by the National Heart, Lung, and Blood Institute and the National Institute on Deafness and Communication Disorders. MACS data collection is also supported by UL1-TR001079 (Johns Hopkins Institute for Clinical and Translational Research, JHU ICTR) from the NCATS, a component of the NIH, and NIH Roadmap for Medical Research. The contents of this publication are solely the responsibility of the authors and do not represent the official views of the NIH, Johns Hopkins ICTR, or NCATS. The MACS website is located at http://aidscohortstudy.org/. J. A. S. was supported by K01AG048765 from NIA. T. T. B. was supported in part by R01AI093520 and K24AI120834 from the NIAID. K. M. E. was supported in part by K23AG050260 and R01AG054366 from the NIA. This work was also supported by the JHU Center for AIDS Research (1P30AI094189). Mitochondrial DNA haplogrouping in MACS was supported by R21DK101342 from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) to Hulgan and Brown.
Potential conflicts of interest. T. H. and T. T. B. report grants from NIH/NIDDK during the conduct of the study. K. E reports grants from NIH, Gilead, and Merck; nonfinancial support from Theratechnologies; and personal fees from EMD Serono, though not directly related to the submitted work. F. J. P. reports grants and personal fees from Gllead Sciences, Janssen, Merck, and ViiV, though not directly related to the submitted work. All remaining authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Mahy M, Autenrieth CS, Stanecki K, Wynd S. Increasing trends in HIV prevalence among people aged 50 years and older: evidence from estimates and survey data. AIDS 2014; 28(Suppl 4):S453–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Deeks SG, Lewin SR, Havlir DV. The end of AIDS: HIV infection as a chronic disease. Lancet 2013; 382:1525–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Greene M, Covinsky K, Astemborski J, et al. . The relationship of physical performance with HIV disease and mortality. AIDS 2014; 28:2711–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Palella FJ Jr, Baker RK, Moorman AC, et al. ; HIV Outpatient Study Investigators Mortality in the highly active antiretroviral therapy era: changing causes of death and disease in the HIV outpatient study. J Acquir Immune Defic Syndr 2006; 43:27–34. [DOI] [PubMed] [Google Scholar]
- 5. Desquilbet L, Jacobson LP, Fried LP, et al. ; Multicenter AIDS Cohort Study HIV-1 infection is associated with an earlier occurrence of a phenotype related to frailty. J Gerontol A Biol Sci Med Sci 2007; 62:1279–86. [DOI] [PubMed] [Google Scholar]
- 6. Schrack JA, Althoff KN, Jacobson LP, et al. ; Multicenter AIDS Cohort Study Accelerated longitudinal gait speed decline in HIV-infected older men. J Acquir Immune Defic Syndr 2015; 70:370–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Piggott DA, Varadhan R, Mehta SH, et al. . Frailty, inflammation, and mortality among persons aging with HIV infection and injection drug use. J Gerontol A Biol Sci Med Sci 2015; 70:1542–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Erlandson KM, Schrack JA, Jankowski CM, Brown TT, Campbell TB. Functional impairment, disability, and frailty in adults aging with HIV-infection. Curr HIV/AIDS Rep 2014; 11:279–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Studenski S, Perera S, Patel K, et al. . Gait speed and survival in older adults. JAMA 2011; 305:50–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schrack JA, Simonsick EM, Chaves PH, Ferrucci L. The role of energetic cost in the age-related slowing of gait speed. J Am Geriatr Soc 2012; 60:1811–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Waters RL, Mulroy S. The energy expenditure of normal and pathologic gait. Gait Posture 1999; 9:207–31. [DOI] [PubMed] [Google Scholar]
- 12. Ferrucci L, Bandinelli S, Benvenuti E, et al. . Subsystems contributing to the decline in ability to walk: bridging the gap between epidemiology and geriatric practice in the InCHIANTI study. J Am Geriatr Soc 2000; 48:1618–25. [DOI] [PubMed] [Google Scholar]
- 13. Eoin R, Patrick W. Impact of mitochondrial toxicity of HIV-1 antiretroviral drugs on lipodystrophy and metabolic dysregulation. Curr Pharm Des 2010; 16:3339–51. [DOI] [PubMed] [Google Scholar]
- 14. Cossarizza A, Pinti M, Nasi M, et al. . Increased plasma levels of extracellular mitochondrial DNA during HIV infection: a new role for mitochondrial damage-associated molecular patterns during inflammation. Mitochondrion 2011; 11:750–5. [DOI] [PubMed] [Google Scholar]
- 15. Hebert SL, Marquet-de Rougé P, Lanza IR, et al. . Mitochondrial aging and physical decline: insights from three generations of women. J Gerontol A Biol Sci Med Sci 2015; 70:1409–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kujoth GC, Hiona A, Pugh TD, et al. . Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science 2005; 309:481–4. [DOI] [PubMed] [Google Scholar]
- 17. Santanasto AJ, Coen PM, Glynn NW, et al. . The relationship between mitochondrial function and walking performance in older adults with a wide range of physical function. Exp Gerontol 2016; 81:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moore AZ, Biggs ML, Matteini A, et al. . Polymorphisms in the mitochondrial DNA control region and frailty in older adults. PLoS One 2010; 5:e11069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kloss‐Brandstätter A, Pacher D, Schönherr S, et al. . HaploGrep: a fast and reliable algorithm for automatic classification of mitochondrial DNA haplogroups. Hum Mutat 2011; 32:25–32. [DOI] [PubMed] [Google Scholar]
- 20. Hendrickson SL, Hutcheson HB, Ruiz-Pesini E, et al. . Mitochondrial DNA haplogroups influence AIDS progression. AIDS 2008; 22:2429–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hulgan T, Haas DW, Haines JL, et al. . Mitochondrial haplogroups and peripheral neuropathy during antiretroviral therapy: an adult AIDS clinical trials group study. AIDS 2005; 19:1341–9. [DOI] [PubMed] [Google Scholar]
- 22. Dudley J, Jin S, Hoover D, Metz S, Thackeray R, Chmiel J. The multicenter AIDS cohort study: retention after 9½ years. Am J Epidemiol 1995; 142:323–30. [DOI] [PubMed] [Google Scholar]
- 23. Graham JE, Ostir GV, Kuo YF, Fisher SR, Ottenbacher KJ. Relationship between test methodology and mean velocity in timed walk tests: a review. Arch Phys Med Rehabil 2008; 89:865–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guralnik JM, Ferrucci L, Simonsick EM, Salive ME, Wallace RB. Lower-extremity function in persons over the age of 70 years as a predictor of subsequent disability. N Engl J Med 1995; 332:556–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Montero-Odasso M, Schapira M, Soriano ER, et al. . Gait velocity as a single predictor of adverse events in healthy seniors aged 75 years and older. J Gerontol A Biol Sci Med Sci 2005; 60:1304–9. [DOI] [PubMed] [Google Scholar]
- 26. Waters RL, Lunsford BR, Perry J, Byrd R. Energy-speed relationship of walking: standard tables. J Orthop Res 1988; 6:215–22. [DOI] [PubMed] [Google Scholar]
- 27. Hreljac A. Determinants of the gait transition speed during human locomotion: kinematic factors. J Biomech 1995; 28:669–77. [DOI] [PubMed] [Google Scholar]
- 28. Mugavero MJ, Napravnik S, Cole SR, et al. ; Centers for AIDS Research Network of Integrated Clinical Systems Cohort Study Viremia copy-years predicts mortality among treatment-naive HIV-infected patients initiating antiretroviral therapy. Clin Infect Dis 2011; 53:927–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cole SR, Napravnik S, Mugavero MJ, Lau B, Eron JJ Jr, Saag MS. Copy-years viremia as a measure of cumulative human immunodeficiency virus viral burden. Am J Epidemiol 2010; 171:198–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Birkus G, Hitchcock MJ, Cihlar T. Assessment of mitochondrial toxicity in human cells treated with tenofovir: comparison with other nucleoside reverse transcriptase inhibitors. Antimicrob Agents Chemother 2002; 46:716–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat 2009; 30:E386–94. [DOI] [PubMed] [Google Scholar]
- 32. Jahn-Eimermacher A. Comparison of the Andersen–Gill model with Poisson and negative binomial regression on recurrent event data. Comput Stat Data Anal 2008; 52:4989–97. [Google Scholar]
- 33. Perera S, Mody SH, Woodman RC, Studenski SA. Meaningful change and responsiveness in common physical performance measures in older adults. J Am Geriatr Soc 2006; 54:743–9. [DOI] [PubMed] [Google Scholar]
- 34. Torroni A, Petrozzi M, D’Urbano L, et al. . Haplotype and phylogenetic analyses suggest that one European-specific mtDNA background plays a role in the expression of Leber hereditary optic neuropathy by increasing the penetrance of the primary mutations 11778 and 14484. Am J Hum Genet 1997; 60:1107–21. [PMC free article] [PubMed] [Google Scholar]
- 35. Hudson G, Carelli V, Spruijt L, et al. . Clinical expression of Leber hereditary optic neuropathy is affected by the mitochondrial DNA-haplogroup background. Am J Hum Genet 2007; 81:228–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gómez-Durán A, Pacheu-Grau D, Martínez-Romero I, et al. . Oxidative phosphorylation differences between mitochondrial DNA haplogroups modify the risk of Leber’s hereditary optic neuropathy. Biochim Biophys Acta 2012; 1822:1216–22. [DOI] [PubMed] [Google Scholar]
- 37. Suissa S, Wang Z, Poole J, et al. . Ancient mtDNA genetic variants modulate mtDNA transcription and replication. PLoS Genet 2009; 5:e1000474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tapper DP, Clayton DA. Mechanism of replication of human mitochondrial DNA. Localization of the 5ʹ ends of nascent daughter strands. J Biol Chem 1981; 256:5109–15. [PubMed] [Google Scholar]
- 39. Payne BA, Wilson IJ, Hateley CA, et al. . Mitochondrial aging is accelerated by anti-retroviral therapy through the clonal expansion of mtDNA mutations. Nat Genet 2011; 43:806–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xue QL. The frailty syndrome: definition and natural history. Clin Geriatr Med 2011; 27:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fried LP, Tangen CM, Walston J, et al. ; Cardiovascular Health Study Collaborative Research Group Frailty in older adults: evidence for a phenotype. J Gerontol A Biol Sci Med Sci 2001; 56:M146–56. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.