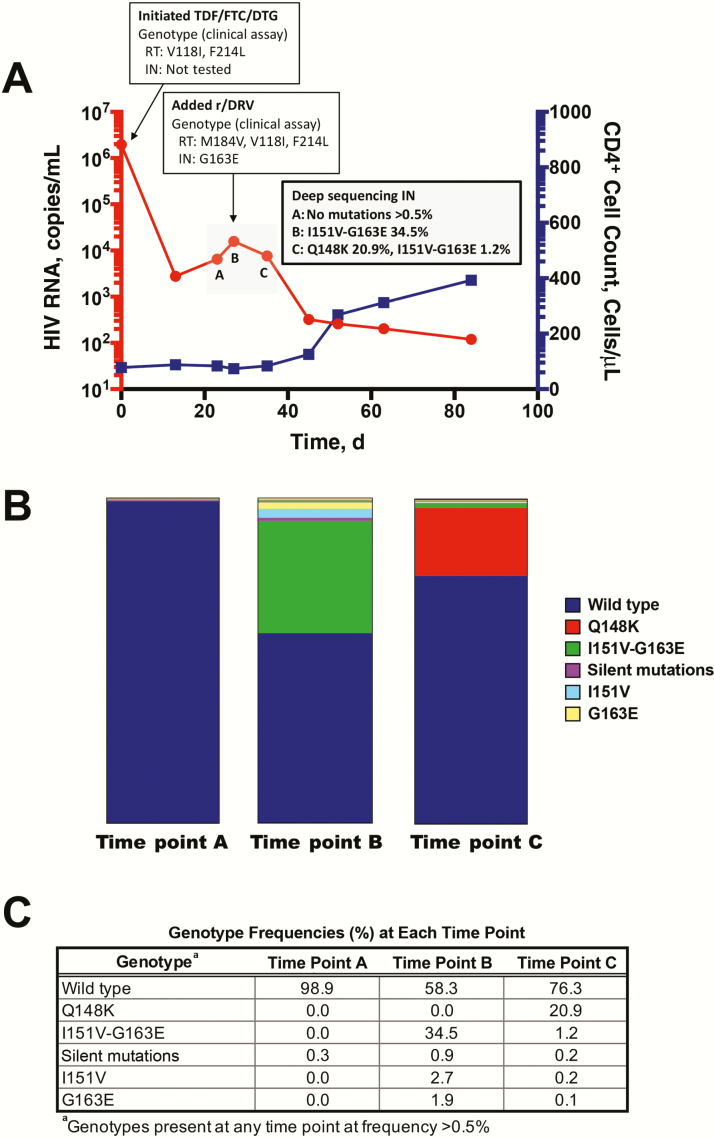
Figure 1.
Rapid development of integrase (IN) strand transfer inhibitor resistance mutations. A, Time course of human immunodeficiency virus type (HIV-1) viremia and CD4+ T lymphocytes. Plasma HIV RNA and absolute CD4+ T-lymphocyte counts from clinical laboratory measurements are plotted from the initiation of antiretroviral therapy (ART). Time points designated A, B, and C indicate those used for paired-end deep sequencing analysis (panel B), and correspond to days 27, 30, and 25 after initiation of ART, respectively. B, Graph of genotypes from IN gene amino acid region 142–165. Deep sequencing analysis was performed on preamplified region of the IN gene from 3 time points during the period of virologic inflection (panel A). Graph shows distribution of genotypes as portion of total reads. C, Complete list of all genotypes from IN gene amino acids 142–165 detected at >0.5% of total reads. Time points refer to those depicted in panel A. Abbreviations: DTG, dolutegravir; FTC, emtricitabine; r/DRV, ritonavir-boosted darunavir; RT, reverse transcriptase; TDF, tenofovir disoproxil fumarate