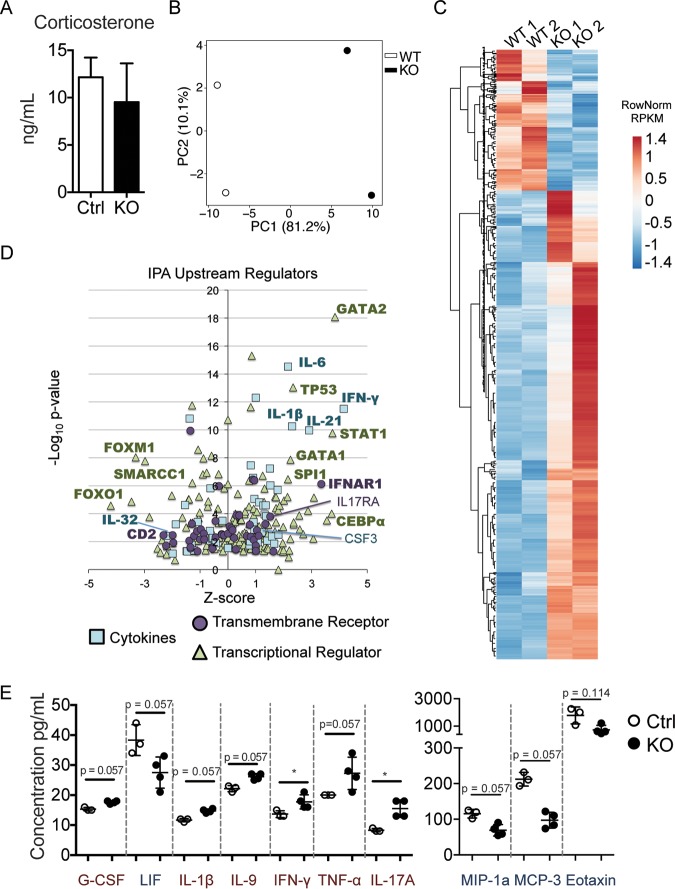
FIG 6.
Zfp521 deficiency affects HSC transcriptomes, potentially through circulating factors within the BM microenvironment. (A) The concentration of serum corticosterone was analyzed by ELISA. Mouse n = 3 littermate controls (Ctrl) and 4 Zfp521−/− knockouts (KO). (B and C) LSK cells were sorted from the BM of KO and Ctrl mice, and RNA was purified and processed for RNA-seq analysis (n = 2 mice each). (B) Principal-component analysis was performed using the top 500 most variable genes. (C) Heatmap showing 947 genes significantly altered (fold change of >2 or <0.5, P value adjusted to <0.1; 728 up and 219 down) in Zfp521−/− LSK relative to Ctrl LSK. (D) IPA analysis of upstream regulator candidates. Cytokines, transmembrane receptors, and transcriptional regulators are shown. Candidates of interest are labeled, and regulators called activated or repressed by IPA are in bold. (E) Soluble protein concentrations were analyzed in BM fluid from Ctrl (n = 3) and KO (n = 4) mice by cytokine array. Trending and significantly upregulated (red) and downregulated (blue) proteins are shown. Statistics were generated using the Mann-Whitney test. *, P < 0.05.