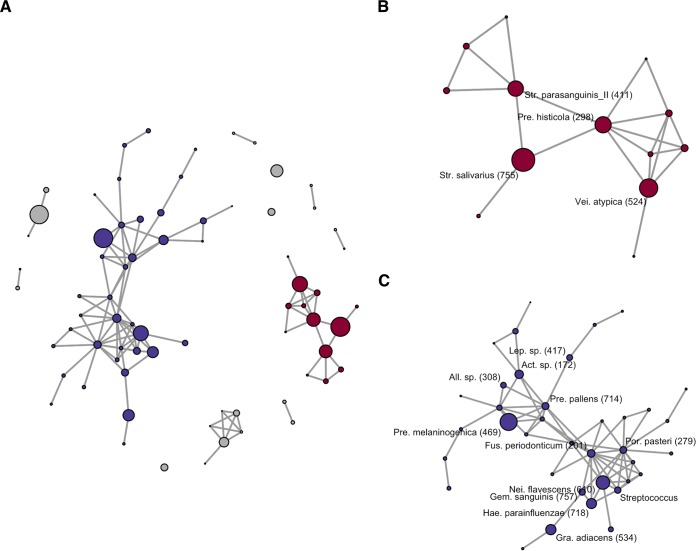
FIG 1 .
Co-occurrence network in tongue microbiota of 506 elderly adults built from SparCC correlation coefficients between sequence abundances. Nodes corresponding to operational taxonomic units (OTUs) and connecting edges indicate the correlation between them. Only correlations with values greater than 0.40 are represented as edges. The node size indicates a mean relative abundance of each OTU. (A) All OTUs excluding isolated nodes corresponding to the OTUs with a mean relative abundance of <1% are shown as nodes in the diagram (72 of all 730 OTUs). Most of the predominant OTUs (with mean relative abundance of >1%) belonged to either of the two major networks including multiple predominant OTUs (cohabiting commensal groups I and II). The nodes belonging to commensal groups I and II are colored red and blue, respectively. (B) Only the co-occurrence network of OTUs belonging to commensal group I is shown. The bacterial taxa corresponding to the predominant OTUs are described. Oral taxon IDs in the Human Oral Microbiome database are given in parentheses following the bacterial names. Abbreviations: Pre., Prevotella; Vei., Veillonella; Str., Streptococcus. (C) Here, only the co-occurrence network of OTUs belonging to commensal group II is shown. The bacterial taxa corresponding to the predominant OTUs are described. Oral taxon IDs in the Human Oral Microbiome database are given in parentheses following bacterial names. Abbreviations: Nei., Neisseria; Hae., Haemophilus; Por., Porphyromonas; Gra., Granulicatella; Fus., Fusobacterium; Pre., Prevotella; Act., Actinomyces; Lep., Leptotorichia; All., Alloprevotella.