Significance
Small-molecule allosteric effectors designed to target and modulate striated and smooth myosin isoforms for the treatment of disease show promise in preclinical and clinical trials. Beta-cardiac myosin is an especially important target, as heart disease remains a primary cause of death in the United States. One prevalent type of heart disease is hypertrophic cardiomyopathy (HCM), which is hypothesized to result from dysregulated force generation by cardiac myosin. Mavacamten is a potent cardiac myosin ATPase activity inhibitor that improves cardiac output in HCM animal models. Our results show that mavacamten selectively stabilizes a two-headed–dependent, autoinhibited state of cardiac myosin in solution. The kinetics and energetics of this state are consistent with the autoinhibited superrelaxed state previously observed only in intact sarcomeres.
Keywords: superrelaxed state, mavacamten, cardiac myosin, allosteric inhibitor, hypertrophic cardiomyopathy
Abstract
We used transient biochemical and structural kinetics to elucidate the molecular mechanism of mavacamten, an allosteric cardiac myosin inhibitor and a prospective treatment for hypertrophic cardiomyopathy. We find that mavacamten stabilizes an autoinhibited state of two-headed cardiac myosin not found in the single-headed S1 myosin motor fragment. We determined this by measuring cardiac myosin actin-activated and actin-independent ATPase and single-ATP turnover kinetics. A two-headed myosin fragment exhibits distinct autoinhibited ATP turnover kinetics compared with a single-headed fragment. Mavacamten enhanced this autoinhibition. It also enhanced autoinhibition of ADP release. Furthermore, actin changes the structure of the autoinhibited state by forcing myosin lever-arm rotation. Mavacamten slows this rotation in two-headed myosin but does not prevent it. We conclude that cardiac myosin is regulated in solution by an interaction between its two heads and propose that mavacamten stabilizes this state.
Familial hypertrophic cardiomyopathies, abbreviated “HCM,” represent one of the most common classes of genetic disease, affecting 1 in 500 people (1). HCM is hypothesized to result from cardiac hypercontractility (2). Direct inhibition of force generation by cardiac myosin is thus a putative treatment (3). Mavacamten (mava), previously termed “Myk461,” is a small-molecule allosteric inhibitor of cardiac myosin that shows promise in preclinical and clinical trials for the treatment of HCM (3). Mava binds with submicromolar affinity to cardiac myosin in the presence of ATP and inhibits steady-state actin-activated and actin-independent (basal) ATPase cycling and calcium-regulated ATPase activity in permeabilized cardiac myofibrils (3, 4). Mava also inhibits the kinetics of rigor actin binding as well as the kinetics of actin-activated phosphate release (4). It decreases in vitro actin filament sliding velocity in the actomyosin motility assay (4), decreases force generation by skinned cardiac preparations from mouse HCM models (3), and decreases cardiac output in live feline hearts (5). Mava’s effect on cardiac contractility is hypothesized to follow from its inhibition of actin-activated phosphate release, the kinetic step most tightly coupled to force generation (4).
Because mava changes the kinetics of actin-activated phosphate release, we hypothesized that it may also stabilize an autoinhibited state of two-headed cardiac myosin in solution, analogous to the superrelaxed (SRX) state observed in skinned myocardium and skeletal muscle fibers (6, 7). This hypothesis was prompted by studies of blebbistatin, a myosin II inhibitor which also inhibits phosphate release and is proposed to stabilize the SRX state in skeletal and cardiac muscle (8–11).
In permeabilized relaxed muscle and myocardium, the SRX state is defined by the biexponential kinetics of single-ATP turnover. The fast phase of this turnover is similar to ATP turnover in isolated single myosin heads (Fig. 1A and SI Appendix, Table S1), while the slow phase is thought to reflect myosin heads that are autoinhibited in a structural state that is folded back onto the filament backbone. The folded state, termed the “interacting heads motif” (IHM), is hypothesized to be stabilized by a number of protein–protein interactions including contacts between the two heads, contacts between the heads and the S2 coiled-coil, and contacts between the heads, the S2 coiled-coil, and the thick-filament backbone. Myosin heads in the IHM are hypothesized to turn over ATP much more slowly than noninteracting heads, because the structural changes in the ATPase site that are required for dissociation of the ATP hydrolysis products require movement of the myosin light-chain–binding domain and actin-binding interface (12, 13), and movement of these elements should be prevented when the heads interact. The IHM is hypothesized to be an evolutionarily conserved autoinhibition mechanism for controlling myosin II driven motility in cells (13–16). EM studies suggest that the individual myosin heads in the cardiac thick filament can form the IHM state (10, 17). However, the IHM has not been observed in purified cardiac myosin in solution without chemical cross-linking. Thus, the structural correlates and transient kinetics of the cardiac myosin IHM state remain unknown.
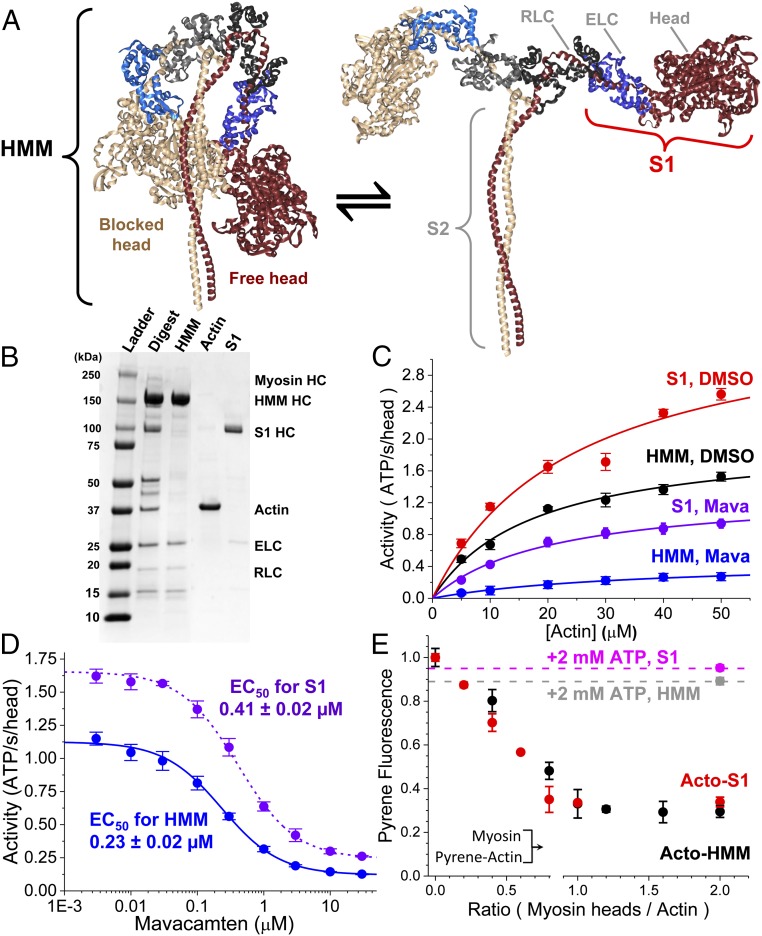
Fig. 1.
Steady-state ATPase activity of purified cardiac myosin fragments HMM and S1. (A) Proposed structural isomerization of HMM between a sequestered IHM [Protein Data Bank (PDB) ID code 5TBY] (Left) and splayed heads (Right). (B) SDS/PAGE gel stained with Coomassie blue demonstrating the purification of α-chymotryptic–digested HMM and S1 myosin fragments, removing contaminating actin. HC, heavy chain of myosin. (C) Steady-state, actin-activated ATPase activity of 0.2 µM S1 with DMSO (red) or 10 μM mava (purple) and of 0.2 μM HMM with DMSO (black) or 10 µM mava (blue). ActivityS1,DMSO = (3.6 ± 0.4⋅s−1) × [Actin]/(24.4 ± 7.4 µM + [Actin]). ActivityHMM,DMSO = (2.02 ± 0.12⋅s−1) × [Actin]/(17.6 ± 2.8 µM + [Actin]). ActivityS1,Mava = (1.35 ± 0.08⋅s−1) × [Actin]/(20.6 ± 3.1 µM + [Actin]). ActivityHMM,Mava = (0.48 ± 0.04⋅s−1) × [Actin]/(35.3 ± 6.3 µM + [Actin]). n = 6 replicates for each actin concentration. (D) Mava is a potent inhibitor of 0.2 µM HMM and S1 during actin-activated, steady-state ATPase cycling; [Actin] = 20 µM. n = 4 replicates. (E) [S1] varied from 0 to 2.0 μM, and [HMM] varied from 0 to 1.0 µM (0- to 2.0-μM heads) mixed with 1.0 µM pyrene-labeled actin. Final concentrations are listed. The HMM and S1 used in this study release from pyrene-labeled actin with the addition of 2.0 mM ATP; FS1 = 0.95 ± 0.1 (magenta) and FHMM = 0.89 ± 0.02 (gray). n = 4 replicates. All experiments were performed in 10 mM Tris (pH 7.5) at 25 °C, 2 mM MgCl2, and 1.0 mM DTT, unless otherwise noted. Error bars indicate SEM.
We tested the hypothesis that cardiac myosin is autoinhibited by direct head–head interaction in solution and that mava selectively targets this intrinsic inhibition by comparing the actin-activated and actin-independent (basal) single-ATP turnover kinetics of single and two-headed bovine ventricular cardiac myosin fragments. Based on the results from those experiments, we evaluated the temperature and ionic strength dependence of ATP turnover by these same myosin preparations. We reasoned that if the two-headed heavy meromyosin (HMM) forms an IHM-driven, autoinhibited state, then the energetics of ATP turnover by HMM should reflect this formation and should be much more dependent on increasing ionic strength than single-headed cardiac myosin S1 (18) and that mava should reduce this dependence on ionic strength. The results from these experiments reveal key aspects of cardiac myosin function and of mava’s mode of action on this critical protein. A related and complementary body of work is described in Anderson et al. (19).
Results
The Steady-State Actin-Activated ATPase kcat of Two-Headed Cardiac Myosin HMM Is Lower than That of Single-Headed Cardiac Myosin Subfragment 1.
We prepared soluble myosin fragments for this study as described in our prior work (20) by isolating cardiac myosin from the left ventricles of bovine hearts, digesting the isolated myosin with α-chymotrypsin (Fig. 1B, lane 2), and purifying the soluble, digested fragments by both size-exclusion and anion-exchange fast protein liquid chromatography as described in SI Appendix. Fractions containing two-headed HMM or the single-headed S1 fragment lacking the regulatory light-chain (RLC)–binding domain and the S2 coiled-coil domain were separated and dialyzed into assay buffers as indicated for each experiment (Fig. 1B, lanes 3 and 5). We verified by gel electrophoresis that the purified HMM and S1 were fully intact and did not contain contaminating cardiac actin (Fig. 1B, lane 4).
We evaluated the functional activity of the purified myosin fragments by measuring actin-activated ATPase activity over a range of actin concentrations using the NADH-coupled ATPase assay (Fig. 1C) (21). Single hyperbolic fits to the actin concentration-dependence of ATPase activity showed that the kcat for the S1 fragment is 3.6 ± 0.4⋅s−1 compared with 2.02 ± 0.12⋅s−1 for cardiac HMM from the identical preparation (P 0.0035) (throughout this paper, measurement statistics and statistical significance are tabulated in SI Appendix, Table S1). Thus, dimerization of the myosin heads reduces the maximum rate of actin-activated ATPase cycling (Fig. 1 A and C).
We verified that the mava preparation (synthesized by EAG Laboratories and verified by mass spectrometry and NMR spectroscopy as described in SI Appendix) inhibits cardiac myosin as demonstrated in prior reports (Fig. 1 C and D) (3). Mava potently inhibited the steady-state ATPase cycling of HMM, decreasing the kcat from 2.02 ± 0.12⋅s−1 to 0.48 ± 0.04⋅s−1 (P 0.001), a 4.21 ± 0.43-fold reduction, and shifting the Km from 17.6 ± 2.8 µM to 35.3 ± 6.3 µM (P 0.05), a twofold increase. The actin-activated ATPase kcat of S1 was inhibited from 3.6 ± 0.4 to 1.35 ± 0.08, a 2.70 ± 0.34-fold reduction. The enhanced inhibition of the actin-activated kcat of HMM compared with S1 is significant (P 0.03). The enhanced inhibition of HMM compared with S1 is reflected in the compounds’ apparent EC50s for inhibition of actin-activated ATPase cycling measured at 20 µM actin (near the Km), which was 0.23 ± 0.02 µM for HMM compared with 0.41 ± 0.02 µM for S1 (P 0.001).
We determined that both purified myosin fragments freely and equivalently bind to actin in the absence of ATP and dissociate from actin upon the addition of 2.0 mM ATP by equilibrating varying concentrations of HMM or S1 myosin with a fixed concentration of pyrene-labeled actin (Fig. 1E). The fluorescence of pyrene-labeled actin is linearly proportional to the binding of individual myosin heads to actin (21). The fluorescence decreased linearly with an increase in the [myosin heads]/[actin] equilibration stoichiometry (Fig. 1E), reaching a maximum at 1:1 stoichiometry with a 70% fluorescence quench, as expected (21). The quench of pyrene fluorescence by HMM or S1 was identical, indicating that both heads of the HMM freely bind actin to a similar degree as S1 heads. The binding is nearly entirely relieved, to 0.89 ± 0.02 or 0.95 ± 0.01, respectively, with the addition of 2.0 mM magnesium ATP (MgATP), and the remaining strongly bound myosin heads are consistent with the expected 5–10% duty-ratio of cardiac myosin estimated in other studies (2). Thus, the S1 and HMM are functional as ATP incubation increases pyrene-actin fluorescence, and the reduced actin activation seen in the HMM does not reflect “dead” or nonfunctional myosin heads which do not detach from actin.
Mava Inhibits Transient Actin-Activated Single-ATP Turnover More for Two-Headed Cardiac HMM than for Single-Headed S1.
We investigated the transient kinetics of actin-activated single-ATP turnover by cardiac myosin HMM and S1 using fluorescently labeled ATP, mant-ATP [2′- or 3′-O-(N-methylanthraniloyl) adenosine 5′-triphosphate]. We mixed the purified myosin fragments by stopped-flow with fluorescent mant-ATP, aged the sample for 2.0 s to allow ATP binding and hydrolysis, and then mixed the resulting steady-state sample with increasing concentrations of actin in the presence of 2.0 mM MgATP. The fluorescence of mant-ATP is enhanced when bound by myosin, and the dissociation of mant-ADP after a single kinetic cycle results in a fluorescent decay (Fig. 2 A and D).
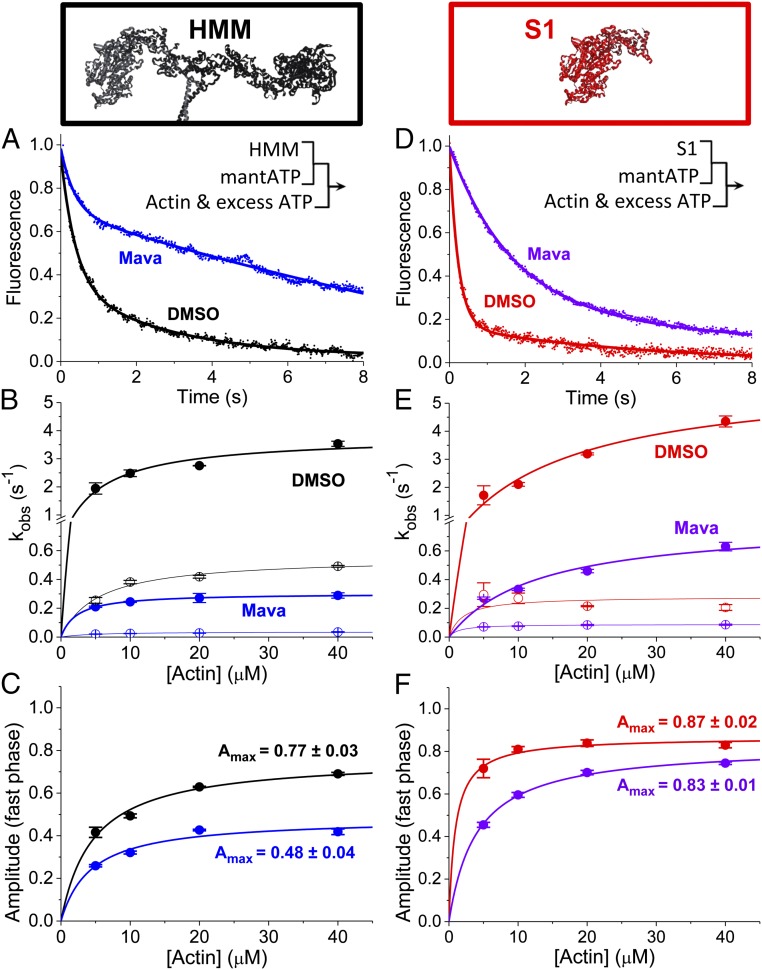
Fig. 2.
HMM data are shown in A–C; S1 data are shown in D–F. (A) Actin-activated single-ATP turnover by 0.1 µM HMM (DMSO control in black, 30 µM mava in blue), mixed with 2.0 μM mant-ATP and 10 μM actin and 2.0 mM MgATP by sequential stopped flow. Premix concentrations are listed. (Inset) Sequential stopped-flow mix schematic. (B and C) Two-exponential fits showing rates (B) and amplitudes (C) with varied [actin]. (D) S1 (0.2 µM) (DMSO in red, 30 µM mava in violet), mixed with 2.0 μM mant-ATP and 10 μM actin and 2.0 mM MgATP by sequential stopped flow. (E and F) Two exponential fits showing rates (E) and amplitudes (F), with varied [actin]. n = 6 replicates, SI Appendix, Table S1. Error bars indicate SEM.
The actin-activated single-ATP turnover fluorescent transients were best fit by a biexponential time-dependent function consistent with previously published studies of actin-activated ATP turnover by cardiac myosin preparations: fluorescence = Afast·exp(−kfast·t) + Aslow·exp(−kslow·t) (22). We evaluated the actin dependence of the two rate constants (kfast and kslow) and the normalized amplitudes (Afast and Aslow) for each phase in the presence or absence of mava (Fig. 2). The fast phase (closed circles in Fig. 2) reflects the release of ATP from a primed, posthydrolysis, myosin.ADP.Pi state ready to undergo actin activation, while the slow phase (open circles in Fig. 2) reflects actin-dependent release from states which are activated more slowly (20, 22). Importantly, the fast and slow phases are both faster than basal single-ATP turnover in the absence of actin (presented in Fig. 3). The maximum observed rate constant for the fast phase of actin-activated ATP turnover was larger for S1 (5.97 ± 0.80⋅s−1) than for HMM (3.80 ± 0.33⋅s−1; P < 0.05). Mava inhibited the maximum observed rate constant for the fast phase of actin-activated ATP turnover to 0.80 ± 0.11⋅s−1 in S1 and 0.30 ± 0.01⋅s−1 in HMM. The maximum observed rate constant for the slow phase was reduced from 0.28 ± 0.04⋅s−1 to 0.089 ± 0.002⋅s−1 in S1 (a threefold inhibition) and from 0.55 ± 0.03⋅s−1 to 0.04 ± 0.01⋅s−1 in HMM (a 14-fold inhibition). Thus, the slow phase of ATP turnover is significantly more inhibited in HMM than in S1 (P 0.0001). In the absence of mava, the amplitude of the slow phase of actin-activated single-ATP turnover is larger in HMM than in S1 (Fig. 2 C and F). At saturating actin concentrations, mava decreased the amplitude of the fast phase in HMM (0.48 ± 0.04) but not in S1 (0.83 ± 0.01; P 0.0001, S1 compared with HMM) (Fig. 2 C and F). The primary difference between S1 and HMM is the presence of the regulatory light chains (RLCs) and the S2 coiled-coil domain and the dimerization of two myosin heads in HMM (Fig. 1A). This dimerization allows the two heads to interact with each other directly and allosterically. Head–head interactions play critical roles in regulating actin activation in other myosin II family members (13). The selective effect of mava on the amplitudes in HMM but not S1 at saturating concentrations of actin indicates that mava stabilizes a state in HMM which occurs infrequently in S1.
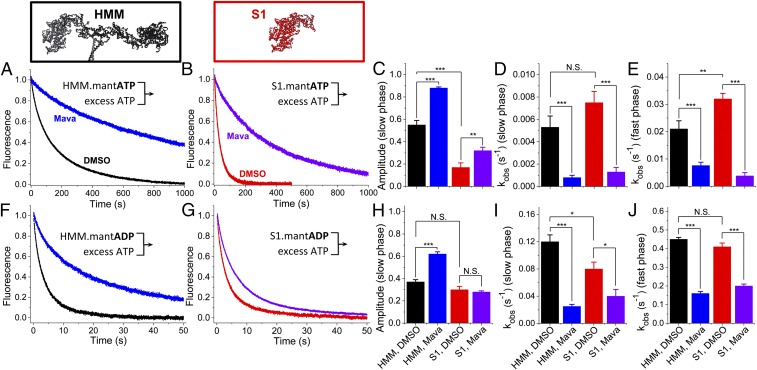
Fig. 3.
Basal nucleotide exchange in the absence of actin. (A and B) Basal single-ATP turnover by stopped-flow mix of 0.2 µM HMM (A) or 0.4 µM S1 (B), mixed with 4.0 μM mant-ATP and then chased with 2.0 mM MgATP. Blue and violet traces indicate 30 µM mava. (C–E) Amplitudes (C) and rates (D and E) of the two-exponential fits of A and B. (F and G) Basal single ADP dissociation from HMM (F) or S1 (G), mixed with 4.0 μM mant-ADP and then chased with 2.0 mM MgATP. (H–J) Amplitudes (H) and rates (I and J) of the two-exponential fits of F and G. n = 9 replicates. Two-exponential fits are reported in SI Appendix, Table S1. Error bars indicate SEM. **P ≤ 0.01, ***P ≤ 0.001; N.S. (not significant), P > 0.05.
Mava Inhibits Basal Single-ATP Turnover and Basal ADP Release in Two-Headed Cardiac HMM Differently than in Single-Headed S1.
We investigated the differences between S1 and HMM in the absence of actin by examining the effects of mava on the kinetics of basal single-ATP turnover and basal ADP release. We mixed 0.4 μM S1 or 0.2 μM HMM—identical concentrations of ATP-binding heads—with either 4.0 μM mant-ATP (Fig. 3 A–E) or 4.0 μM mant-ADP (Fig. 3 F–J) and then measured nucleotide release after mixing with 2.0 mM MgATP. Mava’s effect on ADP release was previously measured in S1 but not in HMM (Fig. 3G) (4).
We analyzed the kinetics of nucleotide exchange in these experiments by fitting the data to a biexponential function. Basal ATP turnover by HMM was distinct from the ATP turnover kinetics measured in S1 (Fig. 3 A–C, black and red traces and bars) exhibiting two distinct kinetic phases. HMM’s slow phase has an amplitude of 0.55 ± 0.04, significantly more than in S1, 0.17 ± 0.04 (Fig. 3C). The rate constants for the fast and slow phase of basal single-ATP turnover are very similar in HMM and S1 (Fig. 3 D, E, I, and J: compare black and red bars). The rate constants we observed for the fast and slow phases of basal single-ATP turnover are 0.02–0.03⋅s−1 and 0.005–0.008⋅s−1, respectively (SI Appendix, Table S1), notably similar to the rate constants for biexponential mant-ATP turnover kinetics detected in permeabilized myocardium where the autoinhibited SRX state has been described (7).
Mava inhibited the rate constants for basal single-ATP turnover in both HMM and S1 preparations to a similar degree (Fig. 3 D, E, I, and J: blue and violet bars). However, as with the actin-activated single-ATP turnover experiments in Fig. 2, the amplitude of the slow phase of basal single-ATP turnover is increased by mava significantly more in HMM than in S1 (Fig. 3C: compare black and blue bars with red and violet bars) (P 0.0001).
The kinetics of ADP release are very similar in HMM and S1 in the absence of mava (Fig. 3 F–J: compare black and red traces and bars). As with ATP turnover, ADP release is inhibited more by mava in HMM (Fig. 3 F and G: compare blue and violet traces). This increased inhibition reflects the stabilization of a slow phase for ADP dissociation that is greatly enhanced from a mole fraction of 0.37 ± 0.02 to 0.62 ± 0.02 (Fig. 3 F and H: compare black and blue traces and bars). The fast and slow rate constants for ADP release are similar in S1 and HMM (Fig. 3 I and J: black and red bars). However, unlike in HMM, mava does not affect the amplitude of the slow phase of ADP dissociation in S1 (Fig. 3H, red and violet bars). These results further support the hypothesis that the structural kinetics of HMM are distinct from those of S1 and that mava stabilizes the slow phase of nucleotide turnover.
The Energetics of Basal Single-ATP Turnover in Two-Headed Cardiac HMM Are Distinct from Those of Single-Headed S1.
We evaluated the biophysical determinants underlying the difference between S1 and HMM described in Fig. 3 by examining the temperature dependence of basal single-ATP turnover in the absence of actin and in the absence of mava. We performed these experiments identically to those in Fig. 3 over a range of temperatures (5.0–35 °C), analyzing the resulting basal single-ATP turnover transients by fitting a biexponential function to the data. The results from these experiments are summarized in Fig. 4. The kinetics of basal single-ATP turnover were distinctly biexponential above 15 °C; we focused our analysis above this temperature (Fig. 3 B, C, E, and F). The rate constants for the fast phase of ATP turnover increased with increasing temperature in both HMM and S1, and the temperature dependence for the increase in kfast and kslow was nearly identical for the two proteins (SI Appendix, Table S1). Fitting to the Eyring equation (given in SI Appendix, Table S1) provides apparent transition-state enthalpy (ΔHǂapp) and a collection of terms associated with the apparent transition-state entropy (ΔSǂapp). For both HMM and S1, the kfast of basal single-ATP turnover has a larger, endothermic ΔHǂapp than the kslow (SI Appendix, Table S1). In addition, for both HMM and S1, kfast has a positive ΔSǂapp, while the kslow has a negative ΔSǂapp. This suggests that kfast and kslow (Fig. 4 B and E) are distinct biophysical processes exhibiting unique transition-state energetics and are rate-limited by unique biochemical or biophysical transitions.
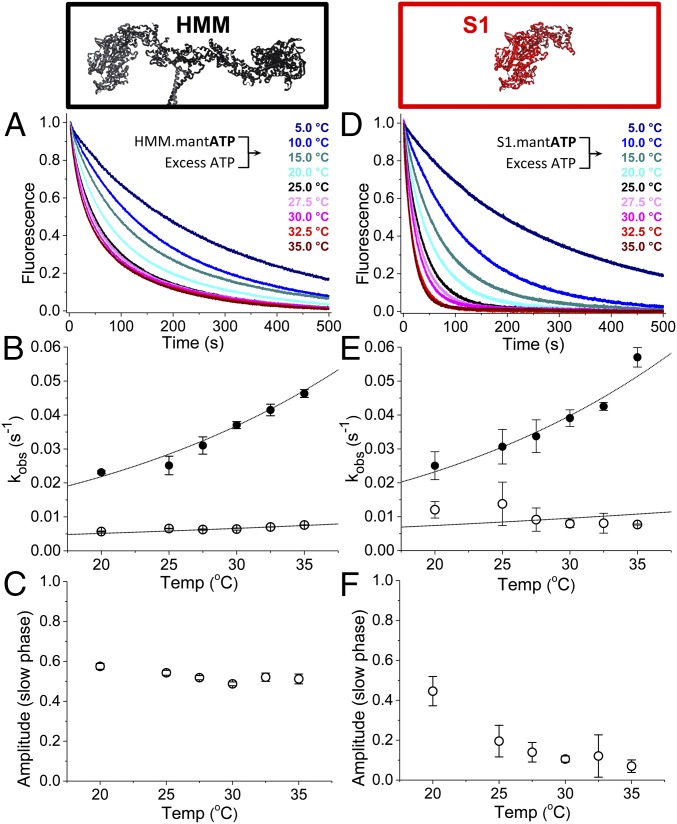
Fig. 4.
Temperature dependence of basal single-ATP turnover. (A and D) 0.4 µM HMM (A) or 0.8 µM S1 (D) mixed with 8.0 µM mant-ATP and then mixed with 2.0 mM MgATP. These data were fit to a two-exponential function. (B) The rates for HMM were fit to the Erying equation: kfast = temperature in Kelvin (T) × exp(−[35.3 kJ/mol]/RT + 5.3)⋅s−1⋅K−1; kslow = T × exp(−[15.9 kJ/mol]/RT − 4.3)⋅s−1⋅K−1. Closed circles indicate the fast phase; open circles indicate the closed phase. (C) The amplitudes of the slow phases depicted in A. (E) The rates from the two-exponential fit for S1 were also fit to the Erying equation: kfast = T × exp(−[30.9 kJ/mol]/RT + 3.6)⋅s−1⋅K−1; kslow = T × exp(−[15.1 kJ/mol]/RT − 3.9)⋅s−1⋅K−1. (F) The amplitudes of the slow phases depicted in D. n = 4 replicates for each temperature. Error bars indicate SEM.
The temperature dependences for the amplitudes of basal single-ATP turnover (Fig. 4 C and F) were also distinct between S1 and HMM samples. In S1, increasing the temperature from 20 to 35 °C dramatically decreased the amplitude of kslow from 0.45 ± 0.07 to 0.07 ± 0.03 (Fig. 4F), while in HMM the amplitude decreased only from 0.57 ± 0.01 to 0.51 ± 0.02 (Fig. 4C). Thus, at near physiologic temperatures (35 °C) the slow phase of basal single-ATP turnover is significantly more abundant in HMM than in S1 (P 0.0001). Temperature stability is consistent with the biochemically sequestered SRX state observed in permeabilized muscle fibers (6) and the ordered state of the myosin thick filament observed by fluorescence polarization (23) and X-ray diffraction (24–26), all of which are stabilized by increasing temperature.
Increasing Ionic Strength Activates Two-Headed Cardiac HMM but Not S1.
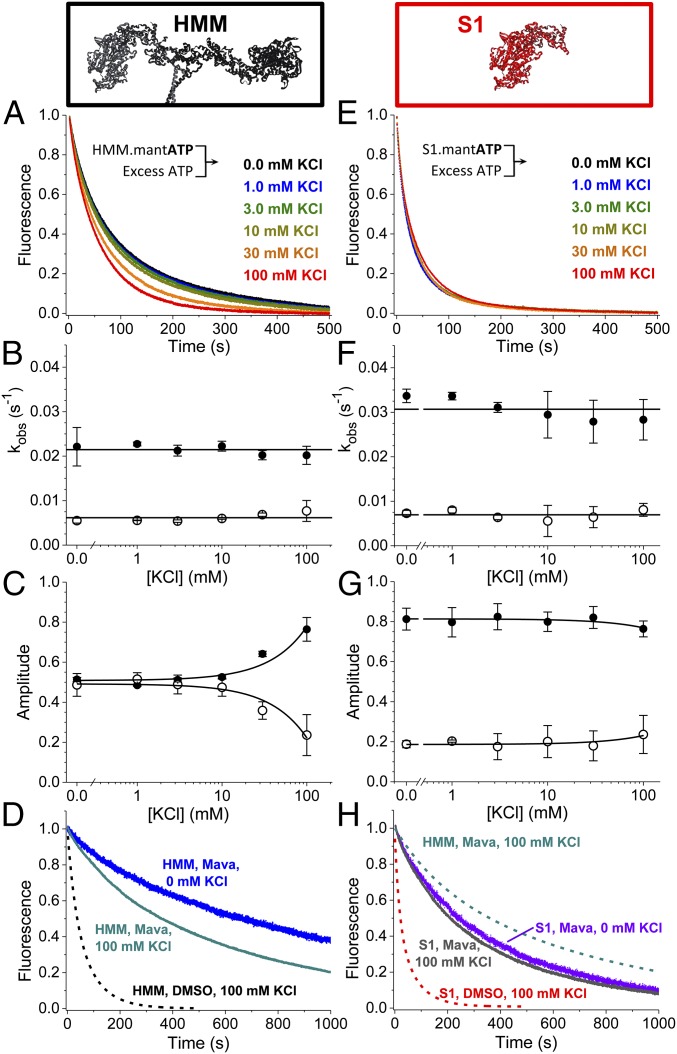
Previous studies showed an isolated S2 coiled-coil domain fragment of human cardiac myosin interacts at low ionic strength with an isolated S1 fragment and with a two-headed HMM truncated at the second heptad of the S2 coiled-coil domain, and an EM study showed the IHM is disrupted in myosin II homologs by increasing ionic strength (13, 18). We therefore examined the ionic strength dependence of basal single-ATP turnover in S1 and HMM at 25 °C by increasing the concentration of potassium chloride (KCl) from 0 mM to 100 mM. The results from these experiments are depicted in Fig. 5. Increasing ionic strength accelerated basal ATP turnover by HMM (Fig. 5 A–C) but had little effect on S1 (Fig. 5 E–G). We fit biexponential functions to the resulting transients. The kfast and kslow rate constants for basal ATP turnover did not change with increasing ionic strength in HMM (Fig. 5B) or in S1 (Fig. 5F). The dependence of turnover by HMM on ionic strength reflected an increase in the amplitude of the fast phase and a decrease in the amplitude of the slow phase, while in S1 the amplitudes did not change. At 100 mM KCl, the amplitude of the fast and slow phases of turnover were similar in HMM and S1 (Fig. 5 C and G). These data suggest that the populations of myosin that turnover ATP fast and slow, exchange in solution, and the energetics of this exchange are dependent on charge–charge interactions.
Fig. 5.
Ionic strength dependence of basal ATP turnover. (A and E) HMM (0.4 µM) (A) or 0.8 µM S1 (E) was mixed with 8.0 µM mant-ATP and then was mixed with 2.0 mM MgATP. Data best fit to two exponentials. (B) The rates of HMM’s basal mant-ATP turnover are relatively constant over these [KCl]; the average values are depicted by the horizontal line. Closed circles represent the fast phase; open circles represent the slow phase. (C) The amplitude of the fast phase increases with increasing [KCl]. Closed circles represent the fast phase; open circles represent the slow phase. Linear fits show trends. (D) In the presence of mava, HMM is sensitive to increasing ionic strength (compare blue and dark cyan traces). (F and G) The rates (F) and amplitudes (G) of S1’s basal mant-ATP turnover are relatively constant. Linear fits show trends. (H) S1 in the presence of mava is insensitive to changes in [KCl] (compare violet and gray traces), consistent with S1 in the absence of mava (E). n = 4 for replicates for each [KCl]. Fits are reported in SI Appendix, Table S1. Error bars indicate SEM.
We also examined the ionic strength dependence of HMM’s autoinhibited state in the presence of mava by performing basal single-ATP turnover under 100-mM KCl or no KCl conditions (Fig. 5D). Increasing the ionic strength to 100 mM KCl partially relieved mava’s inhibition on ATP turnover in HMM but not in S1, indicating that the mava-stabilized state in HMM is specifically dependent on ionic strength (Fig. 5H). Notably, however, at 100 mM KCl and saturating mava, ATP turnover by HMM is still slower and is distinctly biexponential compared with the turnover by S1 (Fig. 5H: compare dashed cyan with gray traces).
Actin Disrupts the Autoinhibited State of Two-Headed Cardiac HMM.
Results in Fig. 3 show that in the absence of actin half of the myosin ATPase sites in HMM turn over ATP more slowly than the sites in S1. This population is stable with increasing temperature (Fig. 4) and is disrupted by increasing ionic strength (Fig. 5), both of which suggest the formation of an auto-inhibitory protein–protein interaction interface. Furthermore, in the presence of mava, the steady-state ATPase kcat (Fig. 1 and SI Appendix, Table S1) and the fast and slow rate constants for actin-activated single-ATP turnover (Fig. 2) are faster than the same kinetics in the absence of actin (Fig. 3). This indicates that actin still activates ATP turnover and ATPase cycling in the presence of saturating mava. Together, these observations suggest that actin interaction relieves the autoinhibition present in HMM even when mava is bound. We reasoned that if the enhanced inhibition of HMM by mava results from a folded, IHM-like structural state, then the structural dynamics of the two light-chain–binding domains of the HMM dimer, which are presumably prevented from moving when HMM is in the IHM state, should be altered, compared with when the HMM is not in the IHM. We tested this hypothesis by using transient time-resolved FRET, (TR)2FRET, to measure these dynamics directly in response to actin activation.
We followed the strategy outlined in our prior work to perform these experiments (20). We labeled the cardiac myosin RLC with the Alexa Fluor-488 fluorescent probe on a single engineered cysteine residue, replacing the native valine at position 105 in the bovine cardiac RLC sequence. We then exchanged the labeled RLC onto purified cardiac HMM to obtain donor-labeled HMM. Expression and purification of the RLC, labeling, and light-chain exchange were performed identically to our prior study and are described in detail in that paper and are outlined in SI Appendix (20). We confirmed that neither the exchange nor the attachment of the fluorescent labels perturbs the autoinhibited state by measuring basal single-ATP turnover (SI Appendix, Fig. S2).
We measured actin-initiated changes in light-chain domain (lever-arm) orientation by equilibrating the donor-labeled HMM with 20-molar excess Cy3-ATP. The Cy3-ATP binds and is hydrolyzed by the myosin. When bound, the Cy3 fluorescent probe is a FRET acceptor for the Alexa-Fluor 488 probe, and thus we obtained FRET-labeled cardiac HMM. FRET between the labeled RLC and the Cy3-ATP reports on the orientation of the myosin light-chain–binding domain (20) [Fig. 1A, bound by the essential light chain (ELC) and RLC]. While binding the Cy3-ATP stabilizes a pre–power-stroke structural state of the light-chain–binding domain, actin stabilizes the post–power-stroke state during the phosphate-release/power-stroke phase and before the dissociation of the hydrolyzed Cy3-ADP. Cy3-ATP stabilization of the pre–power-stroke state is reflected in a decrease in the time-resolved fluorescence lifetime of the Alexa Fluor-488 donor probe, and the power stroke is reflected in the actin-initiated increase in the lifetime (SI Appendix, Fig. S3) (20).
We mixed the Cy3-ATP/Alexa488-HMM complex with increasing concentrations of actin containing a 2 mM excess of MgATP by stopped-flow (Fig. 6A) and acquired time-resolved fluorescence waveforms every 1.0 ms during the resulting actin-induced single-ATP turnover transient (SI Appendix, Fig. S3). We analyzed the changes in the time-resolved fluorescence decay using a two-distance, structure-based model as in our prior study (20). The structural states obtained from fitting this model to the data, summarized in SI Appendix, Table S1, were indistinguishable from our previous results. The center and widths of pre– and post–power-stroke–like structural states were not changed by mava under the conditions of our experiments (SI Appendix, Fig. S5) and therefore were treated as global parameters in the structural model with control samples containing DMSO. We used the same approach in our prior work investigating the effect of omecamtiv mecarbil on the cardiac myosin power stroke (20). From this analysis, we obtain the structural kinetics of the lever-arm rotation in response to actin activation (Fig. 6C and SI Appendix, Table S1).
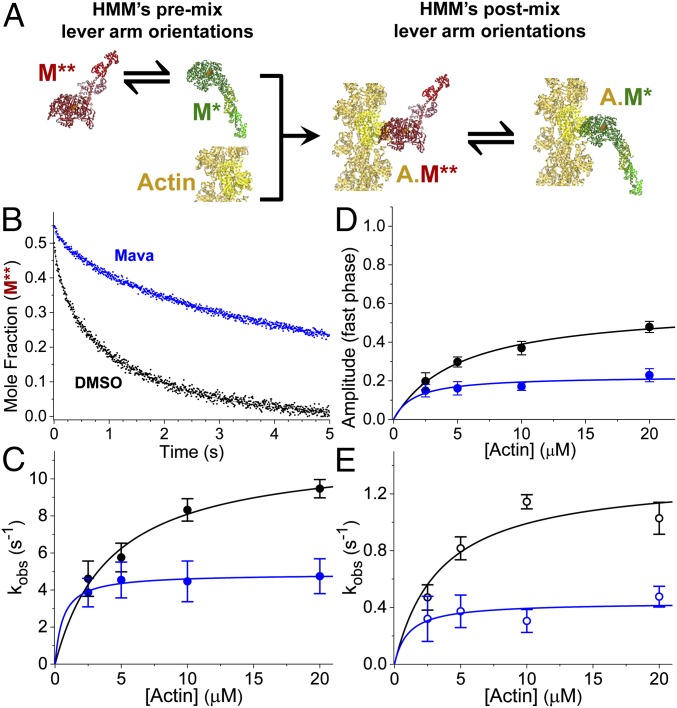
Fig. 6.
Mava inhibits lever-arm rotation in HMM during actin activation as detected with (TR)2FRET. (A) Stopped-flow mix of fluorescently labeled myosin with actin to detect FRET between the lever arm and the catalytic domain of cardiac HMM during the actin-activated power stroke. An equilibrium of structural states for myosin (green or red) are depicted before and after stopped-flow mixing. Fluorophores are located on the RLC and nucleotide. S1 is shown for simplicity; HMM was used. (B) Mole fraction of the M** pre–power-stroke structural state detected with (TR)2FRET. Structural transients are fit to two exponentials. (C) Observed rate constants for the fast phase of the actin-activated power stroke over a range of [Actin]. kobs,fast,DMSO = 11.5⋅s−1[Actin]/(4.2 μM + [Actin]); kobs,fast,Mava = 4.9⋅s−1[Actin]/(0.6 μM + [Actin]). (D) Amplitude of the fast phase: Afast,DMSO = 0.59[Actin]/(5.1 μM + [Actin]); Afast,Mava = 0.23[Actin]/(1.6 μM + [Actin]). (E) Slow phase: kobs.slow,DMSO = 1.3⋅s−1[Actin]/(3.4 μM + [Actin]); kobs,slow,Myk461 = 0.4⋅s−1[Actin]/(1.0 μM + [Actin]). n = 6 replicates of biochemically independent mixes, each averaging 6–10 shots; two separate preparations of cardiac HMM. Error bars indicate SEM.
In the absence of mava, actin drives lever-arm rotation, and the resulting millisecond-timescale mole-fraction transients for each structural state (pre-power stroke short distance or post-power stroke long distance) obtained from fitting the two-state model to the data are best fit to biexponential functions (Fig. 6 C–E and SI Appendix, Fig. S3). The fast phase reflects lever-arm rotation of myosin heads that are primed for activation (Fig. 6C), while the slow phase reflects the equilibration of other HMM species less readily activated by actin (Fig. 6E).
The maximum observed rate constant for the fast phase of actin-activated lever-arm rotation is 11.5 ± 0.8⋅s−1, and the maximum observed rate constant for the slow phase of actin-activated lever-arm rotation is 1.3 ± 0.2⋅s−1. These rate constants are significantly faster than the fast and slow rate constants for actin-activated single-ATP turnover (Fig. 2), indicating that actin binding by the two populations of myosin is followed by FRET-detected lever-arm rotation, followed by ADP release. Saturating mava significantly reduced these rate constants to 4.9 ± 0.2⋅s−1 (P < 0.0001) and 0.43 ± 0.08⋅s−1, respectively (P < 0.001).
We compared the effects of mava on the observed rate constants for actin-activated lever-arm rotation (Fig. 6) and on the rate constants for basal single-ATP turnover (Fig. 3). At saturating mava and actin, the fast phase of the power stroke is 23 ± 3% of the total structural transient and is 16-fold faster than the fast phase of basal ATP turnover in the presence of mava (Figs. 3E and 6 C and D). The slow phase of the power stroke is 77 ± 3% of the transient and is 12-fold faster than the slow phase of basal ATP turnover in the presence of mava (Figs. 3D and 6E). Because both rate constants for the structural change induced by actin are faster than the fastest phase of basal ATP turnover in the absence of mava, we conclude that actin is able to bind the mava-inhibited HMM and to trigger lever-arm rotation in both the fast and slow basal turnover populations. Thus, actin interaction triggers the disruption of the autoinhibited state of cardiac HMM that mava otherwise stabilizes (Fig. 3). We also note that actin activation triggers turnover of all bound Cy3-ATP in the presence of mava, although with slower kinetics than in the absence of the compound. Thus, mava does not permanently trap cardiac myosin in an inhibited state.
Several Steps of the Cardiac Myosin ATPase Cycle Are Not Affected by Mava.
Additional transient stopped-flow kinetics experiments utilizing (TR)2FRET showed that the ATP-induced recovery stroke is not substantially altered by mava (SI Appendix, Fig. S4), nor are ATP binding to myosin or actomyosin, ADP release from actomyosin, myosin dissociation from actomyosin, or the apparent equilibrium constant for ATP hydrolysis measured by acid-quench (SI Appendix, Fig. S6 and Table S1). Mava’s effects on other steps in myosin’s ATPase cycle, such as phosphate release and actin association in the ADP state, have been reported previously (4). We summarize the kinetics steps in the cardiac myosin ATPase cycle that mava inhibits in SI Appendix, Fig. S1 and discuss the implications for these changes below.
Discussion
Regulated activation of the cardiac myosin thick filament is a critical determinant of contraction in the heart. This regulation contributes to the Frank–Starling relationship, a fundamental correlate of cardiac performance (27). It may also contribute to heart diseases (18, 28). Activation of cardiac thick filaments is hypothesized to be controlled by an evolutionarily conserved mechanism through which the heads of each myosin dimer fold back and interact with the dimer’s S2 coiled-coil domain and the underlying surface of the thick filament (13). This structural state, the IHM, has been observed in relaxed muscle (23, 24, 29), in isolated thick filaments, and in purified myosin preparations including glutaraldehyde cross-linked cardiac myosin in the presence of the myosin II inhibitor blebbistatin (11, 13, 30, 31).
Previous muscle-fiber studies revealed a population of myosin ATPase sites with dramatically inhibited ATP-turnover kinetics compared with isolated myosin S1 analyzed in vitro (6, 7, 32). This population is termed the “SRX state.” The SRX biochemical state is hypothesized to result from the formation of the IHM structural state (33); however, their biochemical and structural correlates remain enigmatic. Our results show that an SRX-like biochemical state, indicated by autoinhibited ATP turnover, forms in solution at low ionic strength in the presence of ATP and that this state correlates with changes in the biochemical and structural kinetics of actin activation. This conclusion is based on measured differences between the biochemical kinetics of single-headed cardiac myosin S1 and two-headed HMM.
The steady-state ATPase experiments in Fig. 1 and the transient, actin-activated, single-ATP turnover studies in Fig. 2 demonstrate that key biochemical differences exist between cardiac S1 and HMM. These differences are indicative of head–head–mediated autoinhibition seen in highly regulated two-headed smooth muscle myosins (34, 35) and suggest the compound may target the same allosteric pathway as RLC phosphorylation. In HMM, mava stabilizes the slow phase of ATP turnover in the presence (Fig. 2C) and absence (Fig. 3A) of actin. It also slows actin-independent ADP release to a greater extent in HMM. These changes indicate that interactions between the two heads of the HMM dimer are stabilized by the compound and that this stabilization slows the rate-limiting kinetics of ATP turnover and ADP dissociation (Fig. 3 F–H). We hypothesize that this reflects an increase in the transition-state free energy for opening the phosphate-release pathway and nucleotide-binding pocket in the absence of actin; these transitions limit phosphate and ADP release, respectively. Furthermore, the compound’s lower binding affinity for HMM than for S1, indicated by its EC50 for steady-state ATPase cycling (Fig. 1 and SI Appendix, Table S1), shows that the two-headed myosin adopts a structural state that preferentially binds mava.
The temperature- and ionic strength-dependence studies in Figs. 4 and 5 support the conclusion that the rate-limiting kinetics for ATP turnover in the absence of actin are distinct in HMM and in S1 and reflect the formation of a large protein-interaction interface. The disruption by increasing ionic strength and stability at higher temperatures is consistent with HMM’s two heads interacting in an IHM-like state. Formation of the IHM involves putative ionic interactions (18), and the slow ATP-turnover phase, which we hypothesize correlates with head–head interaction, is disrupted by increasing ionic strength. Stabilization of the slow phase at physiologic temperature is consistent with structural studies in intact muscle that also show slowed ATP turnover and thick-filament ordering at increasing temperature. Importantly, mava stabilizes the autoinhibited state of two-headed HMM in the presence of physiological ionic strengths (Fig. 5 D and H), and thus we expect a similar result in the heart. Recent work suggests this may be the case (19).
Mava changes the structural kinetics of actin activation, consistent with its stabilization of a preexisting autoinhibited state. This conclusion is reflected by the compound’s inhibition of actin-activated lever-arm rotation in Fig. 6. The slowest phase of actin-activated lever arm rotation is significantly faster than the fastest phase of mava-saturated basal ATP turnover by HMM (Fig. 3 A and C–E). This is an important result that indicates actin interaction triggers structural changes in HMM that disrupt the autoinhibited state and that mava binding slows, but does not prevent, these structural changes.
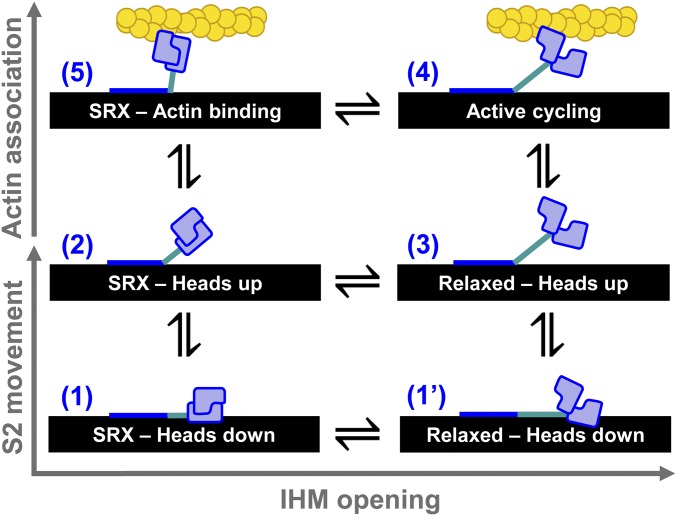
Based on these results, we propose a mechanism for actin activation of the single myosin molecules in the cardiac thick filament (Fig. 7). In this mechanism, autoinhibited myosin heads transition from a folded IHM state that is docked on the thick-filament backbone (state 1) to an IHM state with S2 extended off the filament backbone (state 2). The IHM heads then “open” (state 3) and become available for actin activation (state 4). Thus, myosin in the IHM can be actin-activated by following the pathway state 1 ↔ state 2 ↔ state 3 ↔ state 4 (Fig. 7). Alternatively, actin interaction accelerates IHM head opening if one of the heads—we hypothesize the free IHM head (Fig. 1A) (13)—interacts weakly with available myosin-binding sites on the actin thin filament (state 5 in Fig. 7) and subsequently undergoes a structural transition that disrupts the IHM state. Thus, activation can also proceed via the pathway state 1 ↔ state 2 ↔ state 5 ↔ state 4. This second actin-activated pathway is supported by our biochemical and structural kinetics data and also is depicted in the myosin ATPase cycle in SI Appendix, Fig. S1. The SRX–heads-up state 2 is likely present in muscle fibers because we have observed it in tissue-purified HMM, and it is stable at physiological temperatures (Fig. 4). This state is weakened at physiological ionic strengths (Fig. 5), indicating that additional protein–protein interactions are likely required for its full stabilization in the cardiac sarcomere. Similar arguments were made in a previous paper based on head-to-S2 binding experiments performed by microscale thermophoresis (18). We predict that in muscle fibers mava will shift the apparent equilibrium between the relaxed heads to the IHM heads occupied in states 1 and 2. Recent work shows this may occur (19).
Fig. 7.
Structural model for thick-filament regulation informed by data in this study. (1) EM data support an IHM folded back against the thick filament (black). (1′) The heads may open while the S2 is on the thick filament. (2) Hypothesized state based on our detection of an SRX-like state in HMM. (3) Relaxed myosin with heads splayed and ready to interact with actin. (4) Active actomyosin cycling. (5) Hypothesized state based our detection of actin accelerating the autoinhibited basal ATP turnover of HMM and thus likely unfolding the IHM.
In solution, we propose that mava-saturated HMM follows the pathway through states 1 ↔ 2 ↔ 5 ↔ 4 and that actin accelerates ATP turnover compared with basal turnover. This conclusion is based on the observation that the slowest phases of lever-arm rotation detected by (TR)2FRET are substantially faster than the fastest phase of mava-inhibited ATP turnover in the absence of actin. The transition between state 1 and state 2 is likely to play an important role in muscle. This transition is hypothesized to be controlled by interactions with myosin-binding protein C and passive strain on the thick filament (36, 37). Thus, structural studies in intact muscle will be critical for fully understanding the compound’s mode of action in the heart.
Conclusion
We have detected a biochemically autoinhibited state in tissue-purified cardiac HMM. The kinetics and energetics of this state are consistent with the SRX observed in permeabilized myocardium, and thus we argue that the SRX state can be detected in solution. Mava stabilizes this state, revealing an important aspect of this drug’s mechanism of action that was not previously seen in studies of single-headed myosin. Actin interaction is able to disrupt the autoinhibited state and accelerate the rate-limiting structural transition of head–head splaying that limits ATP turnover in HMM. Thus, weak interaction with actin and cardiac myosin can disrupt autoinhibition, indicating that how the autoinhibited myosin heads are tethered to the thick-filament backbone in muscle is critical. Mechanisms likely regulating this tethering include cardiac myosin RLC phosphorylation (38), binding of myosin-binding protein C to the HMM portion of intact myosin in the thick filament (32), thick-filament mechano-sensing (39), and small molecules (37).
Methods
Steady-State ATPase Activity.
The actin-activated MgATPase activity of purified cardiac myosin HMM or S1 was measured using an NADH-coupled assay (21) performed at 25 °C in 10 mM Tris (pH 7.5) and 2 mM MgCl2 with 1.0 mM DTT. The reaction mix contained 0.2 μM HMM or 0.4 μM S1, varied actin concentrations, and 0.2 mM NADH, 0.5 mM phosphoenol pyruvate, 2.1 mM ATP, 10 U/mL lactic acid dehydrogenase, 40 U/mL pyruvate kinase, and 200 nM HMM. We acquired absorbance at 340 nm every 10 s for 120 s total using a Beckman-Coulter DU640B spectrophotometer.
Transient Kinetics.
Transient biochemical experiments with steady-state fluorescence detection (total fluorescence intensity) were performed on an Applied Photophysics stopped-flow spectrophotometer capable of single-mix and sequential-mix experiments with water-bath temperature control. All experiments were performed at 25 °C unless otherwise stated. The single-mix dead time for this instrument is 1.3 ms. All buffers were filtered and then degassed for 30 min under high vacuum before use.
Single-ATP turnover experiments with mant-ATP were performed with both sequential and single stopped-flow mixes. Samples were excited at 280 nm with a Xe lamp and monochromator and were detected through a 400-nm long-pass filter.
Statistics and Error Analysis.
Individual, representative traces shown throughout the paper depict the average of 6–10 shots of the stopped-flow. All experiments were performed in replicates of n = 4–9 and as biochemically independent experiments. Three separate preparations of cardiac HMM from separate bovine myocardia obtained unfrozen from Pel-Freez Biologicals contributed to the experiments throughout this paper; error bars represent ± SEM. The Student’s t test was used to determine the statistical significance of measured parameters tabulated in SI Appendix, Table S1.
A complete discussion of all methods, including the extraction, digestion, purification, and expression of the proteins utilized in this study, the steady-state and transient kinetics methodologies, and (TR)2FRET data acquisition and analysis is provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank John Lipscomb for use of his sequential stopped-flow instrument, Piyali Guhathakurta, and Sami Chu for experimental assistance, and Lien Phung for helpful discussions. This study was supported by NIH Grants R01AR32961 and R42DA037622 (to D.D.T.) and by American Heart Association Grant 14SDG20480032 (to J.M.M.). J.A.R. was supported by a Graduate Excellence Fellowship from the University of Minnesota. This work was previously deposited in BioRxiv, DOI: https://www.biorxiv.org/content/early/2018/03/23/287425.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1720342115/-/DCSupplemental.
References
- 1.Maron BJ, et al. Prevalence of hypertrophic cardiomyopathy in a general population of young adults. Echocardiographic analysis of 4111 subjects in the CARDIA Study. Coronary Artery Risk Development in (Young) Adults. Circulation. 1995;92:785–789. doi: 10.1161/01.cir.92.4.785. [DOI] [PubMed] [Google Scholar]
- 2.Spudich JA. Hypertrophic and dilated cardiomyopathy: Four decades of basic research on muscle lead to potential therapeutic approaches to these devastating genetic diseases. Biophys J. 2014;106:1236–1249. doi: 10.1016/j.bpj.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Green EM, et al. A small-molecule inhibitor of sarcomere contractility suppresses hypertrophic cardiomyopathy in mice. Science. 2016;351:617–621. doi: 10.1126/science.aad3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kawas RF, et al. A small-molecule modulator of cardiac myosin acts on multiple stages of the myosin chemomechanical cycle. J Biol Chem. 2017;292:16571–16577. doi: 10.1074/jbc.M117.776815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stern JA, et al. A small molecule inhibitor of sarcomere contractility acutely relieves left ventricular outflow tract obstruction in feline hypertrophic cardiomyopathy. PLoS One. 2016;11:e0168407. doi: 10.1371/journal.pone.0168407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stewart MA, Franks-Skiba K, Chen S, Cooke R. Myosin ATP turnover rate is a mechanism involved in thermogenesis in resting skeletal muscle fibers. Proc Natl Acad Sci USA. 2010;107:430–435. doi: 10.1073/pnas.0909468107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hooijman P, Stewart MA, Cooke R. A new state of cardiac myosin with very slow ATP turnover: A potential cardioprotective mechanism in the heart. Biophys J. 2011;100:1969–1976. doi: 10.1016/j.bpj.2011.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilson C, Naber N, Pate E, Cooke R. The myosin inhibitor blebbistatin stabilizes the super-relaxed state in skeletal muscle. Biophys J. 2014;107:1637–1646. doi: 10.1016/j.bpj.2014.07.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu S, White HD, Offer GW, Yu LC. Stabilization of helical order in the thick filaments by blebbistatin: Further evidence of coexisting multiple conformations of myosin. Biophys J. 2009;96:3673–3681. doi: 10.1016/j.bpj.2009.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zoghbi ME, Woodhead JL, Moss RL, Craig R. Three-dimensional structure of vertebrate cardiac muscle myosin filaments. Proc Natl Acad Sci USA. 2008;105:2386–2390. doi: 10.1073/pnas.0708912105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jung HS, Komatsu S, Ikebe M, Craig R. Head-head and head-tail interaction: A general mechanism for switching off myosin II activity in cells. Mol Biol Cell. 2008;19:3234–3242. doi: 10.1091/mbc.E08-02-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gyimesi M, et al. The mechanism of the reverse recovery step, phosphate release, and actin activation of Dictyostelium myosin II. J Biol Chem. 2008;283:8153–8163. doi: 10.1074/jbc.M708863200. [DOI] [PubMed] [Google Scholar]
- 13.Lee KH, et al. Interacting-heads motif has been conserved as a mechanism of myosin II inhibition since before the origin of animals. Proc Natl Acad Sci USA. 2018;115:E1991–E2000. doi: 10.1073/pnas.1715247115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wendt T, Taylor D, Trybus KM, Taylor K. Three-dimensional image reconstruction of dephosphorylated smooth muscle heavy meromyosin reveals asymmetry in the interaction between myosin heads and placement of subfragment 2. Proc Natl Acad Sci USA. 2001;98:4361–4366. doi: 10.1073/pnas.071051098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Craig R, Woodhead JL. Structure and function of myosin filaments. Curr Opin Struct Biol. 2006;16:204–212. doi: 10.1016/j.sbi.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 16.Burgess SA, et al. Structures of smooth muscle myosin and heavy meromyosin in the folded, shutdown state. J Mol Biol. 2007;372:1165–1178. doi: 10.1016/j.jmb.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 17.Al-Khayat HA, Kensler RW, Squire JM, Marston SB, Morris EP. Atomic model of the human cardiac muscle myosin filament. Proc Natl Acad Sci USA. 2013;110:318–323. doi: 10.1073/pnas.1212708110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nag S, et al. The myosin mesa and the basis of hypercontractility caused by hypertrophic cardiomyopathy mutations. Nat Struct Mol Biol. 2017;24:525–533. doi: 10.1038/nsmb.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson RL, et al. Mavacamten stabilizes a folded-back sequestered superrelaxed state of β-cardiac myosin. Proc Natl Acad Sci USA. 2018 doi: 10.1073/pnas.1809540115. [DOI] [Google Scholar]
- 20.Rohde JA, Thomas DD, Muretta JM. Heart failure drug changes the mechanoenzymology of the cardiac myosin powerstroke. Proc Natl Acad Sci USA. 2017;114:E1796–E1804. doi: 10.1073/pnas.1611698114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De La Cruz EM, Ostap EM. Kinetic and equilibrium analysis of the myosin ATPase. Methods Enzymol. 2009;455:157–192. doi: 10.1016/S0076-6879(08)04206-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, White HD, Belknap B, Winkelmann DA, Forgacs E. Omecamtiv mecarbil modulates the kinetic and motile properties of porcine β-cardiac myosin. Biochemistry. 2015;54:1963–1975. doi: 10.1021/bi5015166. [DOI] [PubMed] [Google Scholar]
- 23.Fusi L, Huang Z, Irving M. The conformation of myosin heads in relaxed skeletal muscle: Implications for myosin-based regulation. Biophys J. 2015;109:783–792. doi: 10.1016/j.bpj.2015.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huxley HE, Brown W. The low-angle x-ray diagram of vertebrate striated muscle and its behaviour during contraction and rigor. J Mol Biol. 1967;30:383–434. doi: 10.1016/s0022-2836(67)80046-9. [DOI] [PubMed] [Google Scholar]
- 25.Xu S, Offer G, Gu J, White HD, Yu LC. Temperature and ligand dependence of conformation and helical order in myosin filaments. Biochemistry. 2003;42:390–401. doi: 10.1021/bi026085t. [DOI] [PubMed] [Google Scholar]
- 26.Reconditi M, et al. Sarcomere-length dependence of myosin filament structure in skeletal muscle fibres of the frog. J Physiol. 2014;592:1119–1137. doi: 10.1113/jphysiol.2013.267849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kampourakis T, Ponnam S, Irving M. Hypertrophic cardiomyopathy mutation R58Q in the myosin regulatory light chain perturbs thick filament-based regulation in cardiac muscle. J Mol Cell Cardiol. 2018;117:72–81. doi: 10.1016/j.yjmcc.2018.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trivedi DV, Adhikari AS, Sarkar SS, Ruppel KM, Spudich JA. Hypertrophic cardiomyopathy and the myosin mesa: Viewing an old disease in a new light. Biophys Rev. 2018;10:27–48. doi: 10.1007/s12551-017-0274-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reconditi M, et al. Motion of myosin head domains during activation and force development in skeletal muscle. Proc Natl Acad Sci USA. 2011;108:7236–7240. doi: 10.1073/pnas.1018330108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woodhead JL, et al. Atomic model of a myosin filament in the relaxed state. Nature. 2005;436:1195–1199. doi: 10.1038/nature03920. [DOI] [PubMed] [Google Scholar]
- 31.Woodhead JL, Zhao FQ, Craig R. Structural basis of the relaxed state of a Ca2+-regulated myosin filament and its evolutionary implications. Proc Natl Acad Sci USA. 2013;110:8561–8566. doi: 10.1073/pnas.1218462110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McNamara JW, et al. MYBPC3 mutations are associated with a reduced super-relaxed state in patients with hypertrophic cardiomyopathy. PLoS One. 2017;12:e0180064. doi: 10.1371/journal.pone.0180064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cooke R. The role of the myosin ATPase activity in adaptive thermogenesis by skeletal muscle. Biophys Rev. 2011;3:33–45. doi: 10.1007/s12551-011-0044-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trybus KM, Freyzon Y, Faust LZ, Sweeney HL. Spare the rod, spoil the regulation: Necessity for a myosin rod. Proc Natl Acad Sci USA. 1997;94:48–52. doi: 10.1073/pnas.94.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ellison PA, Sellers JR, Cremo CR. Kinetics of smooth muscle heavy meromyosin with one thiophosphorylated head. J Biol Chem. 2000;275:15142–15151. doi: 10.1074/jbc.275.20.15142. [DOI] [PubMed] [Google Scholar]
- 36.Zhang X, et al. Distinct contributions of the thin and thick filaments to length-dependent activation in heart muscle. eLife. 2017;6:e24081. doi: 10.7554/eLife.24081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kampourakis T, Zhang X, Sun YB, Irving M. Omecamtiv mercabil and blebbistatin modulate cardiac contractility by perturbing the regulatory state of the myosin filament. J Physiol. 2018;596:31–46. doi: 10.1113/JP275050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naber N, Cooke R, Pate E. Slow myosin ATP turnover in the super-relaxed state in tarantula muscle. J Mol Biol. 2011;411:943–950. doi: 10.1016/j.jmb.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fusi L, Brunello E, Yan Z, Irving M. Thick filament mechano-sensing is a calcium-independent regulatory mechanism in skeletal muscle. Nat Commun. 2016;7:13281. doi: 10.1038/ncomms13281. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.