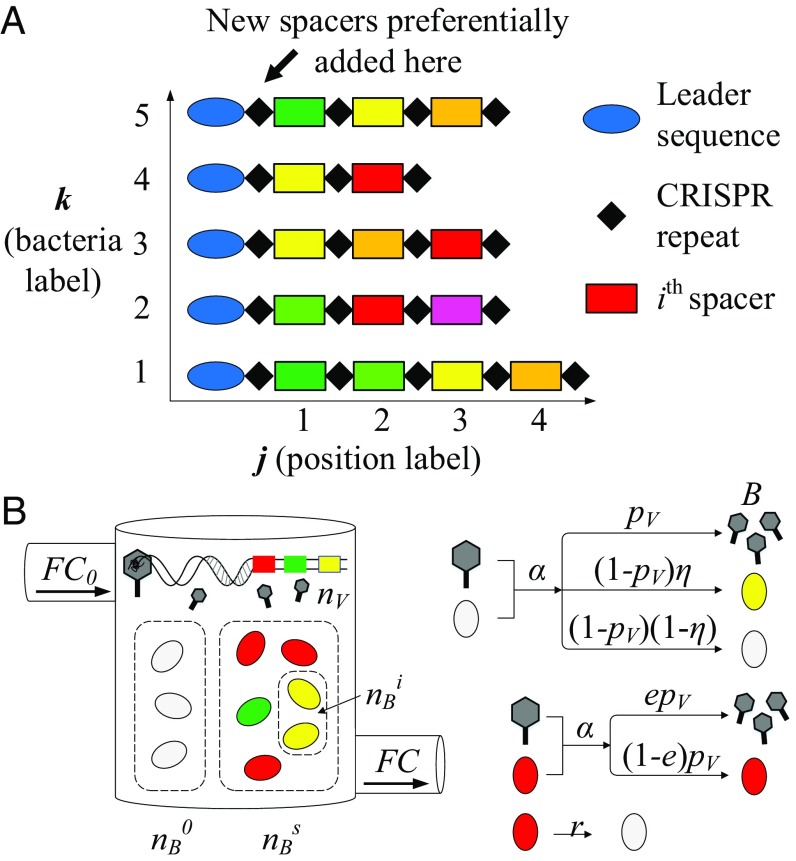
Fig. 1.
(A) CRISPR locus. Small (30 nt) samples of invasive phage DNA called spacers (colored rectangles) are incorporated into the CRISPR genetic locus. Spacers are separated by short (30 nt) sequences called repeats (black diamonds). Multiple spacers can be stored at a CRISPR locus, resulting in a genetic record of immunization (42). In our analysis of the experimental data shown in Fig. 3 A–C, we identify spacers with a type , a locus position , and a bacterium . (B) In our model, bacteria and phages interact in a chemostat (flow cell) with a constant inflow and outflow rate . Nutrients flow into the chemostat at a fixed concentration . Phages are assumed to be identical with a large, fixed number of possible protospacers. Phages adsorb to bacteria with rate and successfully infect and kill naive bacteria with probability . Each bacterium can acquire a single spacer (). Spacers are tracked in the population as the number of bacteria containing a spacer of type , . If a naive bacterium survives an infection, it can acquire a spacer with probability . All spacers are assumed to be equally effective: The probability of phage success in an infection is reduced by if a bacterium has a spacer. Bacteria with spacers revert to naive bacteria by losing a spacer with rate .