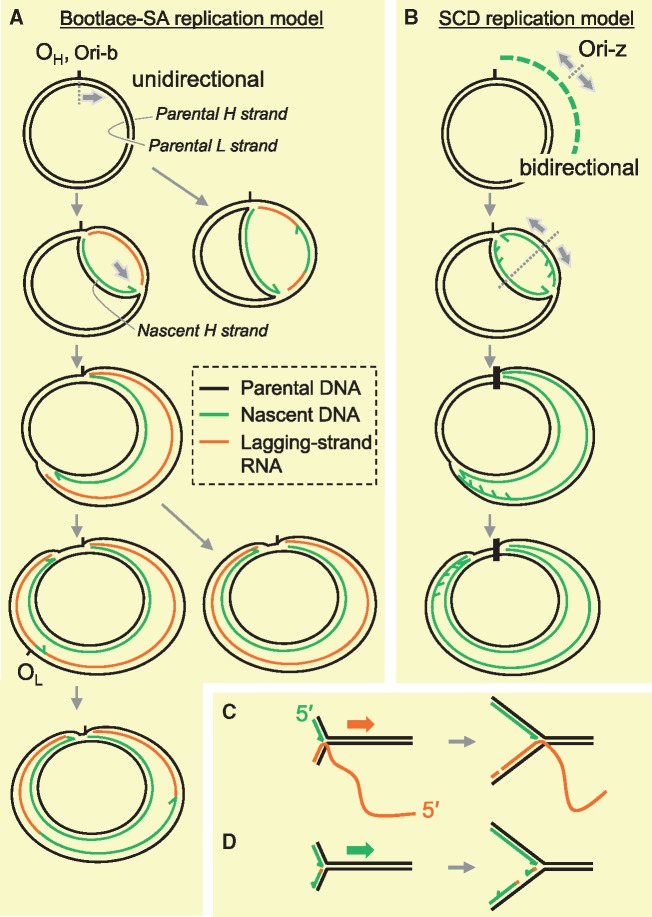
Fig. 4.
Proposed models of mtDNA replication. (A, C) Bootlace strand-asynchronous replication (Bootlace-SA replication). Replication of this mode initiates with the synthesis of an H-strand at one of two sites, OH or Ori-b. H-strand DNA synthesis (leading-strand synthesis) proceeds unidirectionally with concurrent incorporation of RNA into the lagging strand. The RNA lagging strand entails hybridization (‘threading’) of processed mitochondrial transcripts to the parental H-strand, as illustrated in (C). Delayed L-strand DNA synthesis is initiated frequently, but not exclusively at OL in mammals and proceeds unidirectionally. RNA lagging strands are removed in this process. (B, D) Strand-coupled DNA replication (SCD replication). Most of the replication within this mechanism initiates from a broad zone of several kilobases, including the gene-encoding region of mtDNA (Ori-z). In this replication mode, replication is bidirectional and the OH region appears to function as a replication fork barrier. Characterization of RIs from this process suggests that syntheses of the leading and lagging strands are synchronous (coupled) and both strands are essentially composed of DNA. The mechanism of lagging-strand synthesis remains to be elucidated. A proposed model is that lagging strands are formed with multiple short DNA fragments (Okazaki fragments) which are primed by an as yet unidentified mitochondrial primase as illustrated in (D), similar to nuclear DNA replication.