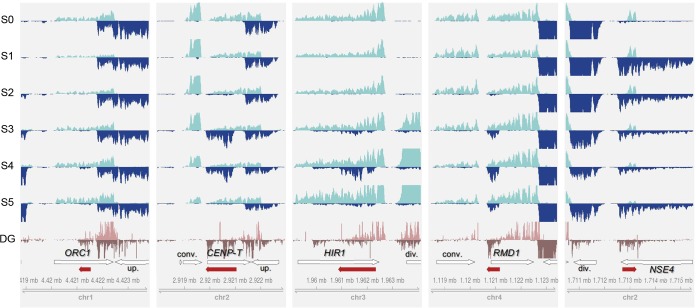
FIG 4 .
Examples of lncRNA expression across the sexual development stages. Per-base coverage of transcripts was plotted for both DNA strands in a 5-kb window. For the perithecial transcriptome data sets (S0 to S5), mapped reads of 3 biological replicates were pooled and then subsampled to 60 million reads for visual comparison of expression levels across the stages. For the degradome-seq data sets (DG), mapped reads of 2 replicates were pooled and displayed. The positions of lncRNAs (red arrows) and their neighboring genes (white arrows) are shown in the annotation track with the genome coordinate at the bottom of each panel. The genes overlapping lncRNAs on the opposite strand are labeled with abbreviated gene names in boldface: CENP-T, centromere protein T (FGRRES_16954), HIR1, histone regulatory protein 1 (FGRRES_05344); NSE4, nonstructural maintenance of chromosome element 4 (FGRRES_17018); ORC1, origin recognition complex subunit 1 (FGRRES_01336); RMD1, required for meiotic division 1 (FGRRES_06759). In relation to lncRNA position, neighboring genes are also labeled as follows: div., divergently transcribed gene on the opposite strand; conv., convergently transcribed gene on the opposite strand; up., upstream gene in tandem on the same strand.