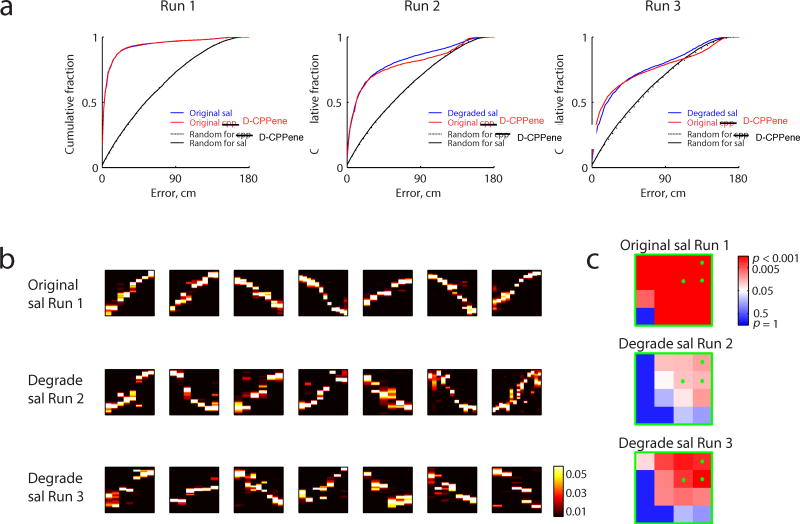
Figure 5. Trajectory Events Under Degraded Saline Place Field Decoder.
(a) Position decoding for each Run using original place fields for D-CPPene days (red) and original/degraded place fields for saline days (blue). For the pre-drug epoch, saline position decoding matches that of D-CPPene and so is included without degradation, as a baseline comparison. For subsequent epochs, noise has been added to the saline epochs until the position decoding matches that of the D-CPPene group. Black and dashed black lines are the decoding qualities at chance level for saline or D-CPPene days. Median ± s.e.m of construction error, Wilcoxon rank sum test: 4.34 ± 0.25 versus 4.86± 0.25, original saline versus D-CPPene in Run 1, p=0.08; 12.68 ± 0.40 versus 13.23± 0.43, saline versus D-CPPene in Run 2, p=0.42; 20.28 ± 0.44 versus 16.94± 0.46, degraded saline versus D-CPPene in Run 3, p<10-10. All the decoding error curves are significantly better than chance (p<10-10). (b) Top seven correlated events expressed during saline Runs using original or degraded saline place field decoder. (c) Corresponding significance matrix for trajectory events.