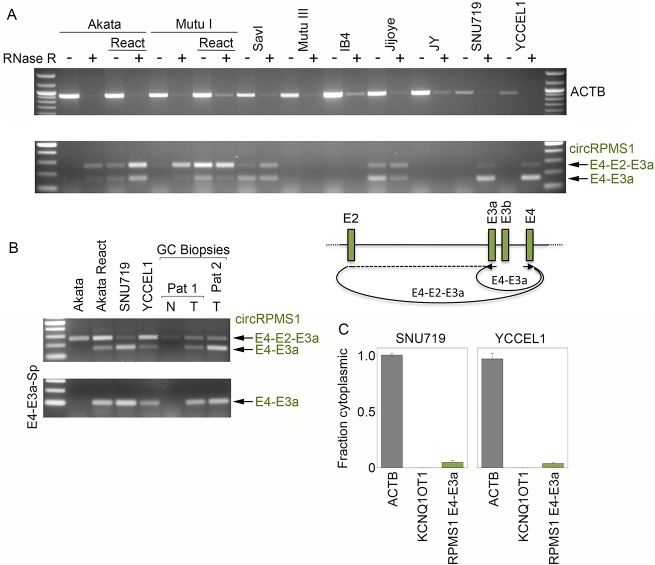
Fig 6. RT-PCR analysis of circRPMS1_E4_E3a and circRPMS1_E4_E2 circular RNAs.
A) RNase R resistance of circRPMS1_E4_E3a and circRPMS1_E4_E2. Divergent primers within RPMS1 exons 4 and 3a detect both exon 4 to exon 3a and exon 4 to exon 2 (exon 4 to exon 2 to exon 3a) circular RNAs (validated by sequencing of excised fragments). All PCR reactions were repeated with similar results. B) circRPMS1_E4_E3a and circRPMS1_E4_E2 are detected in stomach cancer biopsies from two patients (Pat 1 and Pat 2). “N” refers to normal adjacent tissue and “T” refers to tumor tissue. All PCR reactions were repeated with similar results. C) RT-qPCR analysis using junction specific primers shows nuclear enrichment for circRPMS1_E4_E3a. Error bars represent standard deviations and were derived from triplicate qPCR reactions. These experiments were repeated once with similar results. Method for calculating fraction cytoplasmic is outlined in the methods section.