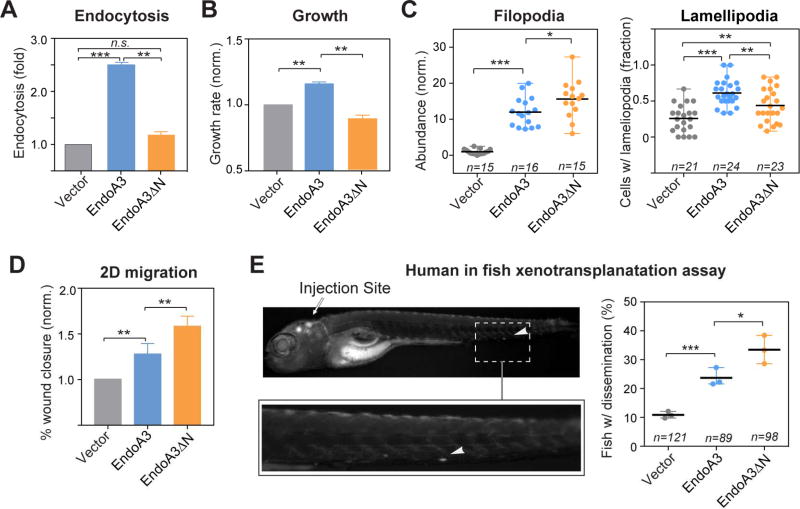
Figure 4. EndoA3ΔN lacking the membrane-binding activity has increased activity in promoting migration.
The comparison of (A) endocytosis, and (B) growth among cells expressing EndoA3ΔN-mCherry (orange), EndoA3WT-mCherry (blue), and control vectors (gray). Error bars indicate SEM. (C) The number of filopodia protrusions per cell is shown in the left panel, and the fraction of cells with lamellipodia is shown in the right panel. “n” = number of images quantified. (D) Wound closure of EndoA3ΔN DLD1 cells was accelerated in the 2D migration assay. Error bars indicate SEM. (E) Xenotransplantation of human cancer cells into zebrafish hindbrain for assessing cell dissemination in vivo. (Upper left panel) A representative image of a zebrafish that was injected with DLD1 cells expressing EndoA3WT-mCherry into the fish embryo hindbrain. The image was taken 96 hours after injection. The white arrowhead indicates DLD1 cells that have disseminated from the injection site (indicated by the white arrow) in the zebrafish embryo hindbrain. (Lower left panel) An image showing the zoomed area with disseminated DLD1 cells. (Right panel) Percentage of fishes with disseminated DLD1 cells expressing empty vectors, EndoA3 WT-mCherry, and EndoA3ΔN-mCherry. “n”, the number of animals analyzed over 3 independent experiments. One-way ANOVA tests were used for multiple comparisons followed by unpaired Student’s t-test used for error analysis (* p<0.05, ** p<0.01, *** p<0.001).