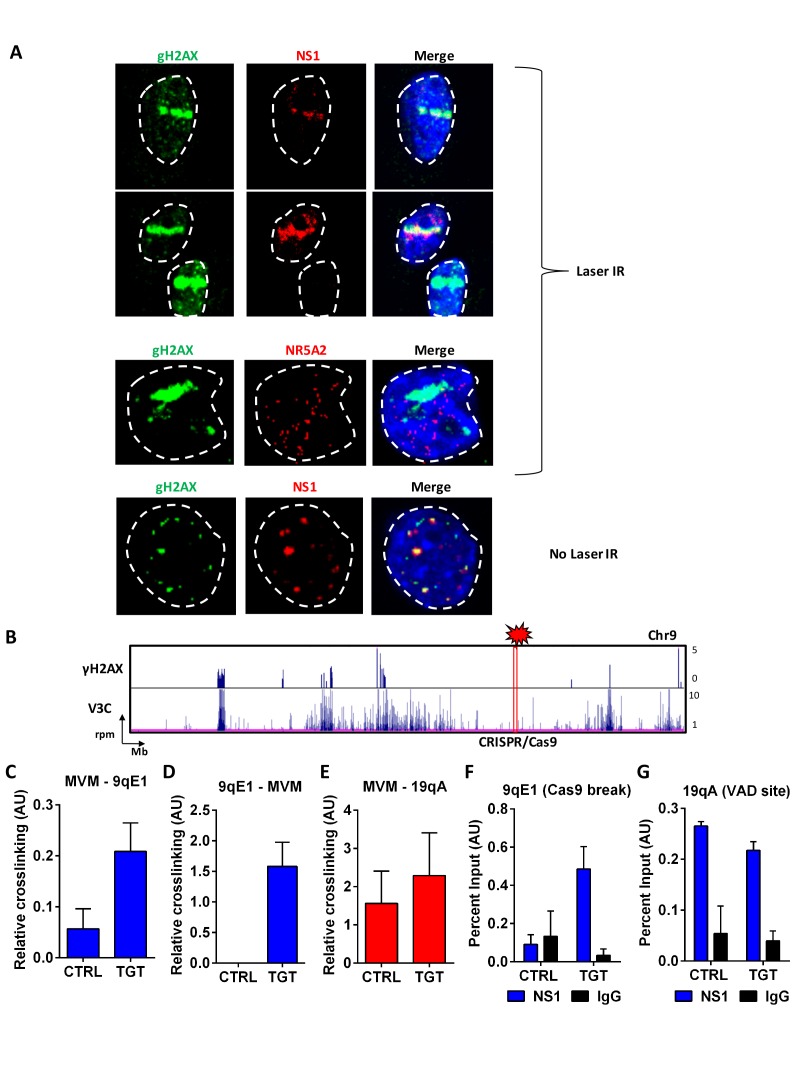
Figure 6. MVM associates with artificially-engineered sites of DNA damage.
(A) Murine A9 fibroblasts were infected with MVM at an MOI of 10 for 18 hr. Cells were sensitized with Hoechst 33342 solution prior to irradiation in selected regions of interest with a 405 nanometer laser (top 3 panels), and were compared with un-irradiated cells (bottom panel). Cells were fixed and analyzed for the localization of DNA damaged sites (detected by green γ-H2AX staining), with that of MVM NS1 protein (red). The irrelevant transcription factor NR5A2 (red) was used as a negative control as this marker that does not colocalize with APAR bodies (see Figure 1). The nuclear periphery is demarcated with dotted white lines and DAPI staining. (B) Schematic of γ-H2AX binding to the cellular genome on chromosome 9 (top panel) compared with MVM-interaction sites (bottom panel) in parasynchronized murine A9 cells infected with MVM (MOI 5) for 16 hr. The site where guide RNAs were designed for DDR induction is shown as a red rectangle and labelled ‘CRISPR/Cas9’. Focused V3C-qPCR assays were performed in A9 cells constitutively expressing LentiCRISPRv2 which were transiently transfected with guide RNAs (nontargeting, labelled as ‘CTRL’ or specific to the 9qE1 site, labelled as ‘TGT’), and infected with MVM at an MOI for 5 for 16 hr. The spatial association of MVM with cellular sites were determined with Taqman probes on (C), the MVM genome, and reciprocal interaction with the Taqman probe on (D), the 9qE1 break site. (E) MVM association with Chr19 VAD at 19qA was tested by focused Taqman qPCR. NS1 occupancy at (F), the 9qE1 break site and (G), the 19qA VAD was assayed by ChIP-qPCR in cells transfected with non-targeting (CTRL) and specific (TGT) guide RNAs. Background levels were determined using IgG pulldowns. qPCR data are presented as mean ± SEM of three independent experiments.