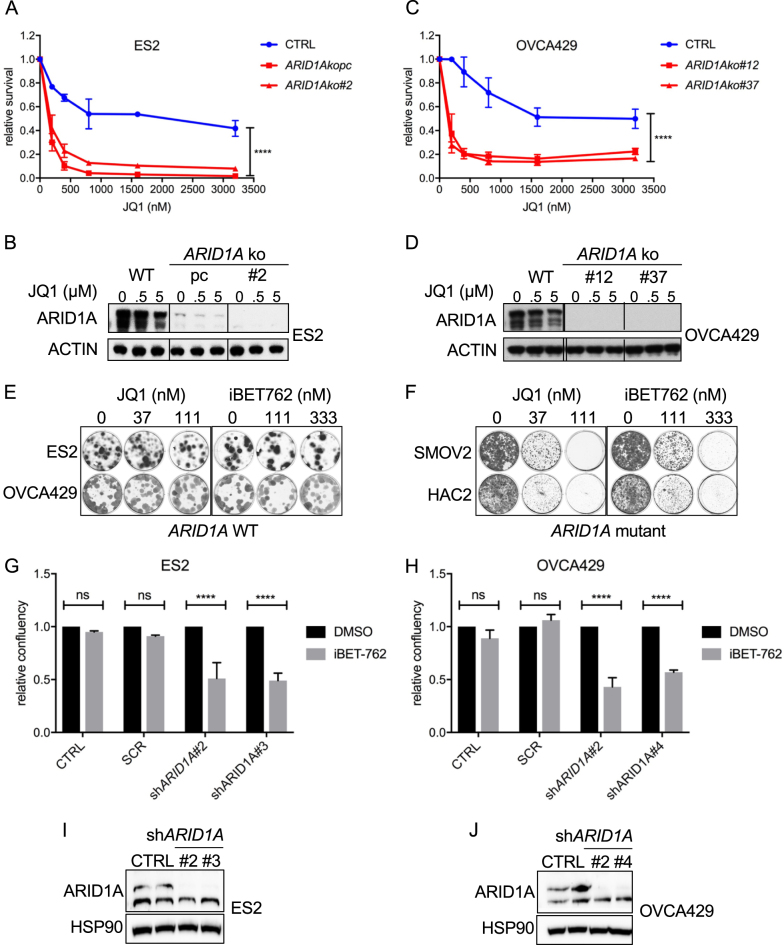
Fig. 3.
ARID1A loss induces sensitivity towards BET inhibitors. a, c ES2 and OVCA429 ARID1A knockout cell lines were generated with a dual vector doxycycline inducible CRISPR/Cas9 vector system. For ES2, wildtype (wt) cells, one polyclonal knockout line (ARID1A kopc) and one monoclonal knockout line (ARID1A ko#2) and for OVCA429, wt cells and two monoclonal knockout lines (ARID1A ko#12 and #37) were tested for JQ1 sensitivity in a 384 well 6-day cell viability assay using CellTiter Blue (CTB) as a readout. CTB measurements were normalized to untreated controls. Error bars denote SD. P-values were calculated with 2-way ANOVA, asterisk denote the number of digits after the decimal. b, d ARID1A Western blot analysis of ARID1A protein levels in the ES2 polyclonal ARID1A knockout clone and ES2/OVCA429 monoclonal ARID1A knockout clones. e, f The indicated two ARID1A wildtype and ARID1A mutant lines were exposed to increasing doses of BET inhibitors JQ1 and iBET762. After 10–12 days cells were stained and photographed. g, h ES2 and OVCA429 cell lines were stably infected with the indicated shRNA constructs targeting ARID1A and plated in 384 well plates. Confluency was monitored in the absence and presence of the BET inhibitor iBET762 (800 nM). Shown are Incucyte confluency measurements relative to untreated cells from three independent experiments. Error bars denote SD. P-values were calculated with multiple t-tests, asterisk denote the number of digits after the decimal. i, j Western blots were performed on the ES2 and OVCA429 cell lines stably infected with the indicated shARID1A constructs to monitor efficiency of ARID1A knockdown. HSP90 served as a loading control