Abstract
Human papillomavirus (HPV) infection alone is not sufficient to explain the development of cervical cancer. Genetic variants have been linked to the development of precancerous lesions and cervical cancer. In this study, we aimed to evaluate the association of 10 single nucleotide polymorphisms (SNPs) of the Fas cell surface death receptor (FAS), trinucleotide repeat containing 6C (TNRC6C), transmembrane channel like 8 (TMC8), DNA meiotic recombinase 1 (DMC1), deoxyuridine triphosphatase (DUT), sulfatase 1 (SULF1), 2′-5-oligoadenylate synthetase 3 (OAS3), general transcription factor IIH subunit 4 (GTF2H4) and interferon gamma (IFNG) genes with susceptibility to precancerous lesions and cervical cancer. In total, 608 female participants, consisting of 199 patients with persistent low-grade precancerous lesions (CIN1), 100 with high-grade precancerous lesions (CIN2/3), 17 patients with cervical cancer and 292 healthy controls, were enrolled in this study. SNPs were tested for associations with each of the above-mentioned cervical group lesions or when considering an overall patient group. A significant difference for rs4737999 was observed between the controls and the overall patient group considering the recessive mode of inheritance [odds ratio (OR), 0.48; 95% confidence interval (CI), 0.24–0.96; P=0.033]. This effect was even stronger on the risk of CIN1 lesions. Carriers of the rs4737999 AA genotype were almost 3-fold less likely of having low grade lesions compared to the other genotypes. On the whole, this study provides evidence of an influence of the SULF1 gene rs4737999 SNP in the development of precancerous lesions/cervical cancer.
Keywords: sulfatase 1, DNA repair, viral infection, cell entry genes, cervical cancer
Introduction
Cervical cancer is estimated to be the fourth most frequent type of cancer among women worldwide (1) and the fourth leading cause of cancer-related mortality (1–3). Environmental factors, living habits and human papillomavirus (HPV) infection have been linked to the development of cervical cancer (4–6). Specifically, HPV infection is considered to be one of the most important causal factors related to cervical cancer (7). However, HPV alone, appears to not be sufficient for the development of cervical cancer (8), as only a small amount of HPV-infected women finally develop cervical cancer (3,9).
A number studies have provided evidence of familial clustering of cervical cancer, supporting the existence of genetic effects (10–12). Moreover, several association studies have demonstrated a number of genetic variants that possibly confer susceptibility to cervical cancer by affecting immune responses, DNA repair or viral cell entry and infection (3,13,14). However, uncertainty for the effect size of genetic variants, particularly in different ethnic backgrounds still exists, as the results of different genetic studies have been conflicting (13).
Recently, Wang et al, genotyped 7,140 SNPs across 305 genes that were involved in HPV infection, cell entry and DNA repair, and reported that genes, among which general transcription factor IIH subunit 4 (GTF2H4), deoxyuridine triphosphatase (DUT), DNA meiotic recombinase 1 (DMC1), 2′-5-oligoadenylate synthetase 3 (OAS3), sulfatase 1 (SULF1), interferon gamma (IFNG), transmembrane channel like 6 (TMC6) and transmembrane channel like 8 (TMC8) were associated with the risk of HPV persistence and cervical pre-cancer/cancer (14). The loss of the expression of DMC1 plays an important role in the development of cancers in human tissues, including cervical cancer lines (15). The DUT enzyme influences nucleotide metabolism by producing the immediate precursor of thymidine nucleotides, dUMP, and consequently decreasing the intracellular concentration of dUTP (16). As a result, uracil cannot be incorporated into DNA (16). SULF1 is a heparin-degrading endosulfatase, which desulfates heparan sulfate proteoglycans (HSPGs) and blocks the binding of growth factors and their receptors, inhibiting as a result, the activation of growth factors and signaling pathways (17,18). OAS3 is induced during viral infection and plays an important role on the antiviral intracellular innate immune response (19). GTF2H4 is a general transcription factor that interacts with factors important in carcinogenesis and is involved in processes of DNA repair and transcriptional control (20). IFNG is regulatory cytokine, released by lymphocytes, that enhances cellular immune responses via increased T-cell cytotoxicity and natural killer (NK)-cell activity (21). The TMC6 and TMC8 genes (also referred to as EVER1 and EVER2 genes), are known for the development of Epidermodysplasia verruciformis, which is associated with a high sensitivity to HPV infections (22). The TMC6 and TMC8 proteins appear to regulate cellular zinc homeostasis in keratinocytes and lymphocyte (23).
In a previous analysis in a population from Northern Greece (3), we failed to detect a significant effect of two SNPs of the EVER1/2 gene region (rs2290907 and rs16970849) and the FAS-670 polymorphism (rs1800682) on precancerous lesions and cervical cancer. This was in contrast to a previous positive study by Castro et al (24). FAS belongs to the family of tumor necrosis factor (TNF) receptors (25,26). The downregulation of FAS leads to resistance to death signals, a phenomenon that has been observed in cervical cancer (27–30).
The present study was designed to replicate the findings reported by Wang et al (14) and Castro et al (24) in a different, from our previous study (3), Greek population of Central Greece. In particular, we examined the effects of 10 SNPs (rs1800682, rs5757133, rs3784621, rs4737999, rs12302655, rs2894054, rs11177074, rs2290907, rs9893818 and [FAS, DMC1, DUT, SULF1, OAS3, GTF2H4, IFNG, TMC6 and TMC8 (2 SNPs)] on the risk of precancerous lesions and cervical cancer.
Materials and methods
Study population
A total of 608 women that had attended the Obstetrics and Gynaecology Clinic of the University Hospital of Larissa, Larissa, Greece participated in this study. The patient group consisted of 316 women with a histopathologically confirmed diagnosis of cervical cancer (n=17) or precancerous lesions, either high grade (CIN2/3, n=100) or persistent low grade (CIN1, n=199). The control group consisted of 292 age-matched women with normal annual cervical cytology screening.
The local Ethics Review Board of the University Hospital of Larissa approved the study protocol. Informed consent was obtained from all individual participants included in the study.
Isolation of DNA and genotyping
Genomic DNA was extracted from 200 µl of EDTA-anti-coagulated whole blood, using a QIAamp® DNA Blood Mini kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer's instructions. SNPs were genotyped with TaqMan allele-specific PCR amplification technology on an ABI PRISM 7900 Sequence Detection System and analyzed with the Sequence Detection Software (SDS 2.1) (both from Applied Biosystems, Foster City, CA, USA) by laboratory personnel blinded to clinical status. In order to assess genotyping reproducibility, initially observed SNP allelic discrimination curves of all genotypes were confirmed by direct DNA sequencing on an ABI PRISM 3100 genetic analyzer (Applied Biosystems).
Statistical analysis
Hardy-Weinberg equilibrium was examined with the exact test Power calculation analysis performed using the CaTS Power Calculator (31). Genotype-disease association analysis was performed with binary logistic regression using the SNPStats platform (http://bioinfo.iconcologia.net/SNPstats/) (32). Odds ratios (ORs), 95% confidence intervals (CIs) and P-values were calculated assuming the co-dominant (genotypic) model (AA vs. Ab vs. bb) and the recessive (AA + Ab vs. bb) modes of inheritance. Four phenotypic groups were searched for the association with the analyzed SNPs compared to the healthy controls: i) The cervical cancer group; ii) the group of patients with high-grade precancerous lesions (CIN2/3); iii) the group of patients with low-grade precancerous lesions (CIN1); and iv) an overall patient group with abnormal cervical changes (either cervical cancer or any type of precancerous lesions).
Results
The characteristics of the 10 studied SNPs (gene, chromosome, chromosomal position, minor allele and minor allele frequencies) are presented in Table I. The genotype call rate was ≥98.85%. All studied SNPs were found to follow the Hardy-Weinberg equilibrium either in the cases or the controls (exact test, P>0.01) (33). Genotype call rate and P-value (exact test) for HWE, for each SNP, are presented and Table II.
Table I.
Characteristics of SNPs genotyped in the current study.
| SNP | rs number | Gene | Chromosome | Chromosome position | Minor allele | MAF CEU | MAF in our control group |
|---|---|---|---|---|---|---|---|
| 1 | rs1800682 | FAS | 10 | 90739943 | C | 0.45 | 0.44 |
| 2 | rs2290907 | TNRC6C | 17 | 76093677 | C | 0.29 | 0.16 |
| 3 | rs16970849 | TMC8 | 17 | 73645503 | A | 0.14 | 0.04 |
| 4 | rs5757133 | DMC1 | 22 | 37277781 | T | 0.23 | 0.31 |
| 5 | rs3784621 | DUT | 15 | 46420384 | C | 0.46 | 0.20 |
| 6 | rs4737999 | SULF1 | 8 | 70680589 | A | 0.20 | 0.27 |
| 7 | rs9893818 | TMC8/TMC6 | 17 | 73653762 | A | 0.04 | 0.00 |
| 8 | rs12302655 | OAS3 | 12 | 111858889 | A | 0.13 | 0.00 |
| 9 | rs2894054 | GTF2H4 | 6 | 30980253 | T | 0.09 | 0.12 |
| 10 | rs11177074 | IFNG | 12 | 66830701 | C | 0.14 | 0.07 |
SNP, single nucleotide polymorphism; MAF, minor allele frequency; CEU, Utah residents with Northern and Western European ancestry; FAS, Fas cell surface death receptor; TNRC6C, trinucleotide repeat containing 6C; TMC8, transmembrane channel like 8; DMC1, DNA meiotic recombinase 1; DUT, deoxyuridine triphosphatase; SULF1, sulfatase 1; OAS3, 2–5-oligoadenylate synthetase 3; IFNG, interferon gamma.
Table II.
Genotype call rate and exact test for HWE of each SNP in the current study.
| Exact test (P-value) for HWE | |||||
|---|---|---|---|---|---|
| SNP | rs number | Gene | Genotype call rate (%) | Controls | Cases |
| 1 | rs1800682 | FAS | 99.18 | 0.096 | 1 |
| 2 | rs2290907 | TNRC6C | 99.67 | 0.83 | 0.36 |
| 3 | rs16970849 | TMC8 | 99.34 | 1 | 1 |
| 4 | rs5757133 | DMC1 | 98.85 | 0.14 | 0.11 |
| 5 | rs3784621 | DUT | 99.34 | 0.85 | 0.6 |
| 6 | rs4737999 | SULF1 | 99.34 | 0.29 | 0.12 |
| 7 | rs9893818 | TMC8/TMC6 | 99.34 | NA | NA |
| 8 | rs12302655 | OAS3 | 99.84 | 1 | 1 |
| 9 | rs2894054 | GTF2H4 | 99.67 | 0.089 | 0.78 |
| 10 | rs11177074 | IFNG | 98.85 | 0.38 | 0.71 |
SNP, single nucleotide polymorphism; HWE, Hardy-Weinberg Equilibrium; NA, non-available; FAS, Fas cell surface death receptor; TNRC6C, trinucleotide repeat containing 6C; TMC8, transmembrane channel like 8; DMC1, DNA meiotic recombinase 1; DUT, deoxyuridine triphosphatase; SULF1, sulfatase 1; OAS3, 2–5-oligoadenylate synthetase 3; IFNG, interferon gamma.
The allelic and genotypic frequencies of the studied SNPs in the control and the overall patient group, as well as in the cervical cancer, high-grade precancerous lesion and low-grade precancerous lesion groups are presented in Table III. Of note, as regards rs9893818, all successfully genotyped participants (100%) carried the CC genotype, whereas as regards rs12302655, >99.0% of the participants carried the wild-type genotype.
Table III.
Allelic and genotype frequencies of SNPs in healthy controls and in cases (cervical cancer cases, cases with low grade and with high grade precancerous lesions).
| SNP | Genotypes/alleles | Controls n (%) | All cases n (%) | Cervical cancer n (%) | High-grade precancerous lesions, n (%) | Low-grade precancerous lesions, n (%) |
|---|---|---|---|---|---|---|
| rs1800682 | ||||||
| Genotype | C/C | 63 (22) | 49 (16) | 2 (12) | 13 (13) | 34 (17) |
| T/C | 129 (44) | 150 (48) | 8 (47) | 50 (51) | 92 (47) | |
| T/T | 98 (34) | 114 (36) | 7 (41) | 36 (36) | 71 (36) | |
| Missed | 2 | 3 | 0 | 1 | 2 | |
| Allele | T | 325 (56) | 378 (60) | 22 (65) | 122 (62) | 234 (59) |
| C | 255 (44) | 248 (40) | 12 (35) | 76 (38) | 160 (41) | |
| rs2290907 | ||||||
| Genotype | C/C | 8 (3) | 4 (1) | 1 (6) | 1 (1) | 2 (01) |
| T/C | 79 (27) | 81 (26) | 3 (0.18) | 19 (0.19) | 59 (0.30) | |
| T/T | 204 (70) | 230 (73) | 13 (0.76) | 80 (0.80) | 137 (0.69) | |
| Missed | 1 | 1 | 0 | 0 | 1 | |
| Allele | T | 487 (84) | 541 (86) | 29 (85) | 179 (90) | 333 (84) |
| C | 95 (16) | 89 (14) | 5 (15) | 21 (10) | 63 (16) | |
| rs16970849 | ||||||
| Genotype | G/A | 24 (8) | 30 (10) | 0 (0) | 10 (10) | 20 (10) |
| G/G | 265 (92) | 285 (90) | 17 (100) | 90 (90) | 178 (90) | |
| A/A | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| Missed | 3 | 1 | 0 | 0 | 1 | |
| Allele | G | 554 (96) | 600 (95) | 34 (100) | 190 (95) | 376 (95) |
| A | 24 (4) | 30 (5) | 0 (0) | 10 (5) | 20 (5) | |
| rs5757133 | ||||||
| Genotype | C/C | 142 (49) | 156 (50) | 11 (65) | 45 (45) | 100 (51) |
| C/T | 114 (39) | 120 (39) | 5 (29) | 43 (43) | 72 (37) | |
| T/T | 34 (12) | 35 (0.11) | 1 (6) | 11 (11) | 23 (12) | |
| Missed | 2 | 5 | 0 | 1 | 4 | |
| Allele | C | 398 (69) | 432 (69) | 27 (79) | 133 (67) | 272 (70) |
| T | 182 (31) | 190 (31) | 7 (21) | 65 (33) | 118 (30) | |
| rs3784621 | ||||||
| Genotype | C/C | 12 (4) | 14 (4) | 2 (12) | 3 (3) | 9 (5) |
| T/C | 92 (32) | 97 (31) | 4 (25) | 30 (30) | 63 (32) | |
| T/T | 187 (64) | 202 (65) | 10 (62) | 66 (67) | 126 (64) | |
| Missed | 1 | 3 | 1 | 1 | 1 | |
| Allele | T | 466 (80) | 501(80) | 24 (75) | 162 (82) | 315 (80) |
| C | 116 (20) | 125 (20) | 8 (25) | 36 (18) | 81 (20) | |
| rs4737999 | ||||||
| Genotype | A/A | 24 (8) | 13 (4) | 1 (6) | 6 (6) | 6 (3) |
| G/A | 106 (37) | 125 (40) | 7 (41) | 37 (37) | 81 (41) | |
| G/G | 160 (55) | 176 (56) | 9 (53) | 57 (57) | 110 (56) | |
| Missed | 2 | 2 | 0 | 0 | 2 | |
| Allele | G | 426 (73) | 477 (76) | 25 (74) | 151 (76) | 301 (76) |
| A | 154 (27) | 151 (24) | 9 (26) | 49 (24) | 93 (24) | |
| rs9893818 | ||||||
| Genotype | C/C | 290 (100) | 316 (100) | 17 (100) | 100 (100) | 197 (100) |
| C/A | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| A/A | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| Missed | 2 | 2 | 0 | 0 | 2 | |
| Allele | C | 580 (100) | 628 (100) | 34 (100) | 200 (100) | 394 (100) |
| A | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| rs12302655 | ||||||
| Genotype | G/G | 292 (100) | 313 (99) | 17 (100) | 99 (99) | 197 (99) |
| G/A | 0 (0) | 2 (1) | 0 (0) | 1 (1) | 1 (1) | |
| A/A | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| Missed | 0 | 1 | 0 | 0 | 1 | |
| Allele | G | 584 (100) | 628 (99.7) | 34 (100) | 199 (100) | 395 (100) |
| A | 0 (0) | 2 (0.3) | 0 (0) | 1 (0) | 1 (0) | |
| rs2894054 | ||||||
| Genotype | C/C | 229 (79) | 246 (78) | 17 (100) | 78 (78) | 151 (76) |
| C/T | 54 (19) | 67 (21) | 0 (0) | 22 (22) | 45 (23) | |
| T/T | 7 (02) | 3 (1) | 0 (0) | 0 (0) | 3 (2) | |
| Missed | 2 | 0 | 0 | 0 | 0 | |
| Allele | C | 512 (88) | 559 (88) | 34 (100) | 178 (89) | 347 (87) |
| T | 68 (12) | 73 (12) | 0 (0) | 22 (11) | 51 (13) | |
| rs11177074 | ||||||
| Genotype | C/C | 0 (0) | 1 (0) | 0 (0) | 0 (0) | 1 (1) |
| T/C | 42 (15) | 52 (17) | 3 (18) | 18 (18) | 31 (16) | |
| T/T | 245 (85) | 261 (83) | 14 (82) | 81 (0.82) | 166 (84) | |
| Missed | 5 | 2 | 0 | 1 | 1 | |
| Allele | T | 532 (93) | 574 (91) | 31 (91) | 180 (0.91) | 363 (92) |
| C | 42 (7) | 54 (9) | 3 (9) | 18 (9) | 33 (8) |
SNPs, single nucleotide polymorphisms. The rows indicating the ‘missed’ numbers indicate the number of failed samples (DNA from some participants failed to be genotyped and consequently there were a few missed genotypes). Percentages (%) have been calculated according to the total number of patients in each group.
Power analysis revealed that our study had a statistical power of >80.0% to detect an genetic association with an OR of 1.78, under the assumption of the multiplicative model, a minor allele frequency of 5% (the lowest in cases for the rs16970849), a type I error level of 0.05, in a sample size consisting of 292 controls and 316 cases (data not shown).
Binary logistic regression analysis demonstrated a significant effect of SULF1 rs4737999 on the risk of the abnormal cervical changes. In particular, a significant difference was observed between the controls and the overall patient group (low-grade, high-grade and cervical cancer) considering the recessive mode of inheritance (OR, 0.48; 95% CI, 0.24–0.96; P=0.033). Individuals carrying the AA genotype had almost half a risk of having cervical cancer, and low- or high-grade lesions compared to those carrying either the GG or the GA genotypes. Moreover, this effect was even more potent on the risk of low-grade precancerous lesions (OR, 0.36; 95% CI, 0.14–0.92; P=0.042) and (OR, 0.35; 95% CI, 0.14–0.87; P=0.014) in the co-dominant and recessive models, respectively. Carriers of the rs4737999 AA genotype were almost 3-fold less likely of having low-grade lesions compared to carriers of the other genotypes. No other SNP was found to alter the risk of any examined phenotype (Table IV).
Table IV.
Single locus analysis.
| All cases (n=316) vs. controls (n=292) | Low-grade (n=199) vs. controls (n=292) | High-grade (n=100) vs. controls (n=292) | Cancer (n=17) vs. controls (n=292) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| SNP | Genotype | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value |
| rs1800682 | |||||||||
| Codominant | T/T | 1.00 | 0.16 | 1.00 | 0.47 | 1.00 | 0.15 | 1.00 | 0.54 |
| T/C | 1.00 (0.70–1.43) | 0.98 (0.66–1.48) | 1.06 (0.64–1.74) | 0.87 (0.30–2.48) | |||||
| C/C | 0.67 (0.42–1.06) | 0.74 (0.44–1.25) | 0.56 (0.28–1.14) | 0.44 (0.09–2.21) | |||||
| Recessive | T/T-T/C | 1.00 | 0.056 | 1.00 | 0.22 | 1.00 | 0.054 | 1.00 | 0.84 |
| C/C | 0.67 (0.44–1.01) | 0.75 (0.47–1.19) | 0.54 (0.29–1.04) | 0.48 (0.11–2.16) | |||||
| rs2290907 | |||||||||
| Codominant | T/T | 1.00 | 0.37 | 1.00 | 0.33 | 1.00 | 0.12 | 1.00 | 0.57 |
| T/C | 0.91 (0.63–1.31) | 1.11 (0.74–1.66) | 0.61 (0.35–1.08) | 0.60 (0.17–2.15) | |||||
| C/C | 0.44 (0.13–1.49) | 0.37 (0.08–1.78) | 0.32 (0.04–2.59) | 1.96 (0.23–16.89) | |||||
| Recessive | T/T-T/C | 1.00 | 0.19 | 1.00 | 0.16 | 1.00 | 0.27 | 1.00 | 0.51 |
| C/C | 0.45 (0.14–1.53) | 0.36 (0.08–1.72) | 0.36 (0.04–2.89) | 2.21 (0.26–18.77) | |||||
| rs16970849 | |||||||||
| G/G | 1.00 | 0.6 | 1.00 | 0.5 | 1.00 | 0.61 | 1.00 | 0.091 | |
| G/A | 1.16 (0.66–2.04) | 1.24 (0.67–2.31) | 1.23 (0.56–2.66) | 0.00 (0.00-NA) | |||||
| rs5757133 | |||||||||
| Codominant | C/C | 1.00 | 0.95 | 1.00 | 0.86 | 1.00 | 0.77 | 1.00 | 0.42 |
| T/C | 0.96 (0.68–1.35) | 0.90 (0.61–1.33) | 1.19 (0.73–1.93) | 0.57 (0.19–1.68) | |||||
| T/T | 0.94 (0.55–1.58) | 0.96 (0.53–1.73) | 1.02 (0.48–2.18) | 0.38 (0.05–3.04) | |||||
| Recessive | C/C-T/C | 1.00 | 0.86 | 1.00 | 0.98 | 1.00 | 0.87 | 1.00 | 0.42 |
| T/T | 0.95 (0.58–1.58) | 1.01 (0.57–1.77) | 0.94 (0.46–1.94) | 0.47 (0.06–3.66) | |||||
| rs3784621 | |||||||||
| Codominant | T/T | 1.00 | 0.97 | 1.00 | 0.97 | 1.00 | 0.84 | 1.00 | 0.4 |
| T/C | 0.98 (0.69–1.38) | 1.02 (0.69–1.50) | 0.92 (0.56–1.52) | 0.81 (0.25–2.66) | |||||
| C/C | 1.08 (0.49–2.39) | 1.11 (0.46–2.72) | 0.71 (0.19–2.59) | 3.12 (0.61–15.85) | |||||
| Recessive | T/T-T/C | 1.00 | 0.83 | 1.00 | 0.82 | 1.00 | 0.62 | 1.00 | 0.19 |
| C/C | 1.09 (0.49–2.39) | 1.11 (0.46–2.68) | 0.73 (0.20–2.63) | 3.32 (0.68–16.29) | |||||
| rs4737999 | |||||||||
| Codominant | G/G | 1.00 | 0.096 | 1.00 | 0.042 | 1.00 | 0.75 | 1.00 | 0.89 |
| G/A | 1.07 (0.77–1.50) | 1.11 (0.76–1.62) | 0.98 (0.61–1.59) | 1.17 (0.42–3.25) | |||||
| A/A | 0.49 (0.24–1.00) | 0.36 (0.14–0.92) | 0.70 (0.27–1.80) | 0.74 (0.09–6.11) | |||||
| Recessive | G/G-G/A | 1.00 | 0.033 | 1.00 | 0.014 | 1.00 | 0.45 | 1.00 | 0.71 |
| A/A | 0.48 (0.24–0.96) | 0.35 (0.14–0.87) | 0.71 (0.28–1.78) | 0.69 (0.09–5.45) | |||||
| rs9893818 | NA | NA | NA | NA | |||||
| rs12302655 | |||||||||
| G/G | 1.00 | 0.1 | 1.00 | 0.18 | 1.00 | 0.098 | NA | NA | |
| G/A | NA (0.00-NA) | NA (0.00-NA) | NA (0.00-NA) | NA | |||||
| rs2894054 | |||||||||
| Codominant | C/C | 1.00 | 0.28 | 1.00 | 0.46 | 1.00 | 0.1 | 1.00 | 0.02 |
| T/C | 1.15 (0.77–1.72) | 1.26 (0.81–1.97) | 1.20 (0.68–2.09) | 0.00 (0.00-NA) | |||||
| T/T | 0.40 (0.10–1.56) | 0.65 (0.17–2.55) | 0.00 (0.00-NA) | 0.00 (0.00-NA) | |||||
| Recessive | C/C-T/C | 1.00 | 0.15 | 1.00 | 0.48 | 1.00 | 0.041 | 1.00 | 0.37 |
| T/T | 0.39 (0.10–1.51) | 0.62 (0.16–2.42) | 0.00 (0.00-NA) | 0.00 (0.00-NA) | |||||
| rs11177074 | |||||||||
| Codominant | T/T | 1.00 | 0.42 | 1.00 | 0.39 | 1.00 | 0.41 | 1.00 | 0.74 |
| T/C | 1.16 (0.75–1.81) | 1.09 (0.66–1.80) | 1.30 (0.71–2.38) | 1.25 (0.34–4.54) | |||||
| C/C | NA (0.00-NA) | NA (0.00-NA) | NA | NA | |||||
| Recessive | T/T-T/C | 1.00 | 0.25 | 1.00 | 0.18 | NA | NA | NA | NA |
| C/C | NA (0.00-NA) | NA (0.00-NA) | NA | NA | |||||
SNP, single nucleotide polymorphism; CI, confidence interval; OR, odds ratio. Statistically significant values are shown in bold; NA, not available.
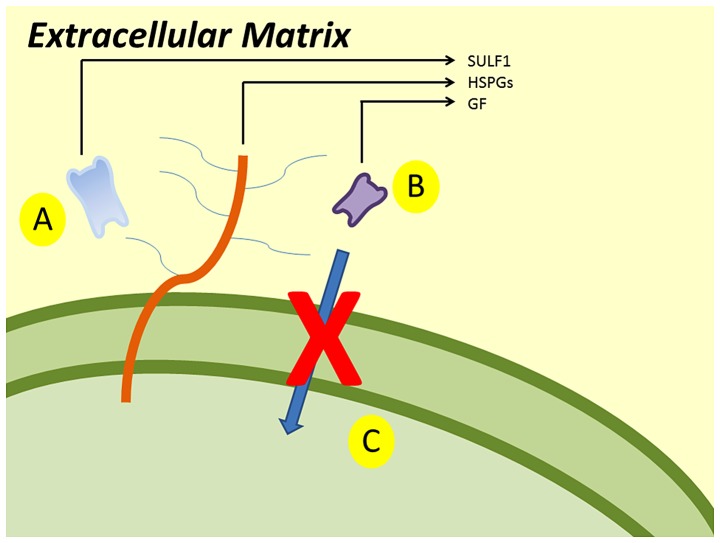
The main mechanism of SULF1 gene is presented in Fig. 1. SULF1 gene encodes the SULF1 protein. SULF1 is a heparin-degrading endosulfatase, which desulfates HSPGs and blocks the binding of growth factors and their receptors. Consequently, it inhibits the activation of growth factor and the signaling pathways.
Figure 1.
SULF1 desulfates HSPGs (A) and blocks the binding of growth factors and their receptors (B). As a result it inhibits the activation of growth factor and the signaling pathways (C).
Discussion
In the present study, we tried to replicate the findings of previous studies regarding the role of SNPs in DNA repair, viral infection and cell entry, and their effects on the risk of cervical cancer and precancerous lesions (14,24). In addition, we re-examined an independent Greek cohort in order to determine the influence of the rs1800682 (FAS), rs2290907 (TMC6) and rs16970849 (TMC8) gene variants (3). In the present study, we found that a specific variant of the SULF1 gene, rs4737999, was associated with a significantly decreased risk of developing precancerous lesions and cervical cancer.
The SULF1 gene is located at the 8q13.3 region. It encodes the homonymous protein, a heparin-degrading endosulfatase, which desulfates HSPGs and blocks the binding of growth factors and their receptors and as a result it inhibits the activation of growth factor and the signaling pathways (Fig. 1) (17,18). The expression of SULF1 appears to be stable in normal tissues, whereas it is downregulated in several tumor cells (34). Moreover, the proliferation and migration of tumor cells can be inhibited by the re-expression of the SULF1 gene (35).
In the study by Wang et al (14), the SULF1 gene reached a statistically significant threshold for the CIN3/Cancer group compared to the controls (P=0.0030). Moreover, the SULF1 gene was also associated with HPV persistence (P=0.005). Additionally, according to SNP-Based association analysis, when the CIN3/Cancer group was compared to the control group, 3 out of the 77 examined SULF1 SNPs (rs4737999, rs4284050 and rs10108002) achieved statistical significance (P-value trend <0.05). In this analysis of Wang et al, the strongest association was reported for the rs4737999; this polymorphism also was associated with precancerous lesions and cervical cancer in the present study.
A number of SNPs of the SULF1 gene have been found to influence the risk of cancer. The AA genotype of r3802278, a SNP located in the 3′-untranslated region (3′-UTR) of the SULF1 gene, was found to play a protective role against breast cancer (36). Moreover, rs2623047, a 5′-upstream gene variant in SULF1, has been associated with an increased risk of breast cancer, as well as with an earlier age of onset and the survival of ovarian cancer (37). Finally, rs6990375, a 3′ prime UTR variant, has been associated with earlier age of ovarian cancer (38). The SNP rs4737999, that reached a statistically significant threshold in the present study, is an intronic non-coding variant located between exons 13 and 14. Therefore, the SULF1 gene may represent an important locus linked to tumorigenesis, as SNPs located in the 5′-upstream region, in the 3′-UTR region or even in the middle of the gene have been found to alter the risk of cancer.
A number of studies have reported that the FAS-670 gene promoter polymorphism is associated with cervical carcinogenesis (39–43). Moreover, the expression of the FAS/FASL genes and the CD95-CD95 ligand (FAS/FASL) interaction seem to confer susceptibility to the development of cervical cancer (3). However, the present study failed to detect any significant effect of the FAS gene SNPs on the risk of cervical cancer or any precancerous lesion. This is in accordance with the results of our previous study in another Greek cohort (3). It is possible that ethnic differences in FAS gene variability may account for the different results among populations (44).
In conclusion, the present study confirms the finidngs of previous reports regarding the role of SULF1 in the risk of precancerous lesions and cervical cancer. This association may have prognostic and pharmacogenetic implications to precancerous lesions or cervical cancer, as SULF1 may be considered as a therapeutic target or biomarker (45,46). Our findings need to be replicated in other populations of other ethnic backgrouns and in experimental models, in order to elucidate the possible role of polymorphic variants of the SULF1 gene in the pathophysiology of mechanism of tumorigenesis.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- SNP
single nucleotide polymorphism
- FAS
Fas cell surface death receptor
- TNRC6C
trinucleotide repeat containing 6C
- TMC8
transmembrane channel like 8
- DMC1
DNA meiotic recombinase 1
- DUT
deoxyuridine triphosphatase
- SULF1
sulfatase 1
- OAS3
2′-5-oligoadenylate synthetase 3
- GTF2H4
general transcription factor IIH subunit 4
- IFNG
interferon gamma
- HPV
human papillomavirus
- TMC6
transmembrane channel like 6
- OR
odds ratio
- CI
confidence interval
- TNF
tumor necrosis factor
- HSPGs
heparan sulfate proteoglycans
- 3′-UTR
3′-untranslated region
- CD95
cluster of differentiation 95
- FASL
Fas ligand
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
Eda and VS were involved in the conceptualization of the study, data curation, formal analysis and investigation, methodology, project administration, resources, software, study supervision, validation, writing of the original draft and writing, reviewing and editing the manuscript. AG, EP, MK, GX, ED, GG and ENK provided the patient blood samples and clinical information, and were involved in data validation, and in the writing, reviewing and editing of the manuscript. DAS and AT were involved in data investigation and validation, as well as in the writing, reviewing and editing of the manuscript. AD was involved in the conceptualization of the study, data curation, formal analysis and investigation, methodology, project administration, resources, software, study supervision, validation, writing of the original draft and writing, reviewing and editing the manuscript. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The local Ethics Review Board of the University Hospital of Larissa approved the study protocol. Informed consent was obtained from all individual participants included in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Pavlidou E, Daponte A, Egea R, Dardiotis E, Hadjigeorgiou GM, Barbadilla A, Agorastos T. Genetic polymorphisms of FAS and EVER genes in a Greek population and their susceptibility to cervical cancer: A case control study. BMC Cancer. 2016;16:923. doi: 10.1186/s12885-016-2960-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schoell WM, Janicek MF, Mirhashemi R. Epidemiology and biology of cervical cancer. Semin Surg Oncol. 1999;16:203–211. doi: 10.1002/(SICI)1098-2388(199904/05)16:3<203::AID-SSU2>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 5.Boda D, Docea AO, Calina D, Ilie MA, Caruntu C, Zurac S, Neagu M, Constantin C, Branisteanu DE, Voiculescu V, et al. Human papilloma virus: Apprehending the link with carcinogenesis and unveiling new research avenues (Review) Int J Oncol. 2018;52:637–655. doi: 10.3892/ijo.2018.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Libra M, Scalisi A, Vella N, Clementi S, Sorio R, Stivala F, Spandidos DA, Mazzarino C. Uterine cervical carcinoma: Role of matrix metalloproteinases (Review) Int J Oncol. 2009;34:897–903. doi: 10.3892/ijo_00000215. [DOI] [PubMed] [Google Scholar]
- 7.Bosch FX, Lorincz A, Muñoz N, Meijer CJLM, Shah KV. The causal relation between human papillomavirus and cervical cancer. J Clin Pathol. 2002;55:244–265. doi: 10.1136/jcp.55.4.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baritaki S, Sifakis S, Huerta-Yepez S, Neonakis IK, Soufla G, Bonavida B, Spandidos DA. Overexpression of VEGF and TGF-beta1 mRNA in Pap smears correlates with progression of cervical intraepithelial neoplasia to cancer: Implication of YY1 in cervical tumorigenesis and HPV infection. Int J Oncol. 2007;31:69–79. [PubMed] [Google Scholar]
- 9.Castellsagué X, Muñoz N. Chapter 3: Cofactors in human papillomavirus carcinogenesis - role of parity, oral contraceptives, and tobacco smoking. J Natl Cancer Inst Monogr. 2003;2003:20–28. doi: 10.1093/oxfordjournals.jncimonographs.a003477. [DOI] [PubMed] [Google Scholar]
- 10.Magnusson PK, Lichtenstein P, Gyllensten UB. Heritability of cervical tumours. Int J Cancer. 2000;88:698–701. doi: 10.1002/1097-0215(20001201)88:5<698::AID-IJC3>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 11.Mammas IN, Zafiropoulos A, Spandidos DA. Involvement of the ras genes in female genital tract cancer. Int J Oncol. 2005;26:1241–1255. [PubMed] [Google Scholar]
- 12.Koffa M, Koumantakis E, Ergazaki M, Malamoumitsi V, Spandidos D. Detection of ras gene-mutations and hpv in lesions of the human female reproductive-tract. Int J Oncol. 1994;5:189–195. [PubMed] [Google Scholar]
- 13.Chen X, Jiang J, Shen H, Hu Z. Genetic susceptibility of cervical cancer. J Biomed Res. 2011;25:155–164. doi: 10.1016/S1674-8301(11)60020-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang SS, Gonzalez P, Yu K, Porras C, Li Q, Safaeian M, Rodriguez AC, Sherman ME, Bratti C, Schiffman M, et al. Common genetic variants and risk for HPV persistence and progression to cervical cancer. PLoS One. 2010;5:e8667. doi: 10.1371/journal.pone.0008667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harada H, Nagai H, Tsuneizumi M, Mikami I, Sugano S, Emi M. Identification of DMC1, a novel gene in the TOC region on 17q25.1 that shows loss of expression in multiple human cancers. J Hum Genet. 2001;46:90–95. doi: 10.1007/s100380170115. [DOI] [PubMed] [Google Scholar]
- 16.Mol CD, Harris JM, McIntosh EM, Tainer JA. Human dUTP pyrophosphatase: Uracil recognition by a beta hairpin and active sites formed by three separate subunits. Structure. 1996;4:1077–1092. doi: 10.1016/S0969-2126(96)00114-1. [DOI] [PubMed] [Google Scholar]
- 17.Ji W, Yang J, Wang D, Cao L, Tan W, Qian H, Sun B, Qian Q, Yin Z, Wu M, et al. hSulf-1 gene exhibits anticancer efficacy through negatively regulating VEGFR-2 signaling in human cancers. PLoS One. 2011;6:e23274. doi: 10.1371/journal.pone.0023274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu H, Fu X, Ji W, Liu K, Bao L, Yan Y, Wu M, Yang J, Su C. Human sulfatase-1 inhibits the migration and proliferation of SMMC-7721 hepatocellular carcinoma cells by downregulating the growth factor signaling. Hepatol Res. 2013;43:516–525. doi: 10.1111/j.1872-034X.2012.01080.x. [DOI] [PubMed] [Google Scholar]
- 19.Thamizhmani R, Vijayachari P. Association of dengue virus infection susceptibility with polymorphisms of 2′-5′-oligoadenylate synthetase genes: a case-control study. Braz J Infect Dis. 2014;18:548–550. doi: 10.1016/j.bjid.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gervais V, Lamour V, Jawhari A, Frindel F, Wasielewski E, Dubaele S, Egly JM, Thierry JC, Kieffer B, Poterszman A. TFIIH contains a PH domain involved in DNA nucleotide excision repair. Nat Struct Mol Biol. 2004;11:616–622. doi: 10.1038/nsmb782. [DOI] [PubMed] [Google Scholar]
- 21.Sun Y, Lu Y, Pen Q, Li T, Xie L, Deng Y, Qin A. Interferon gamma +874 T/A polymorphism increases the risk of cervical cancer: Evidence from a meta-analysis. Tumour Biol. 2015;36:4555–4564. doi: 10.1007/s13277-015-3100-4. [DOI] [PubMed] [Google Scholar]
- 22.Orth G. Host defenses against human papillomaviruses: Lessons from epidermodysplasia verruciformis. Curr Top Microbiol Immunol. 2008;321:59–83. doi: 10.1007/978-3-540-75203-5_3. [DOI] [PubMed] [Google Scholar]
- 23.Lazarczyk M, Dalard C, Hayder M, Dupre L, Pignolet B, Majewski S, Vuillier F, Favre M, Liblau RS. EVER proteins, key elements of the natural anti-human papillomavirus barrier, are regulated upon T-cell activation. PLoS One. 2012;7:e39995. doi: 10.1371/journal.pone.0039995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castro FA, Ivansson EL, Schmitt M, Juko-Pecirep I, Kjellberg L, Hildesheim A, Gyllensten UB, Pawlita M. Contribution of TMC6 and TMC8 (EVER1 and EVER2) variants to cervical cancer susceptibility. Int J Cancer. 2012;130:349–355. doi: 10.1002/ijc.26016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Contreras DN, Krammer PH, Potkul RK, Bu P, Rossi JL, Kaufmann AM, Gissmann L, Qiao L. Cervical cancer cells induce apoptosis of cytotoxic T lymphocytes. J Immunother. 2000;23:67–74. doi: 10.1097/00002371-200001000-00009. [DOI] [PubMed] [Google Scholar]
- 26.Dybikowska A, Sliwinski W, Emerich J, Podhajska AJ. Evaluation of Fas gene promoter polymorphism in cervical cancer patients. Int J Mol Med. 2004;14:475–478. [PubMed] [Google Scholar]
- 27.Butler LM, Hewett PJ, Butler WJ, Cowled PA. Down-regulation of Fas gene expression in colon cancer is not a result of allelic loss or gene rearrangement. Br J Cancer. 1998;77:1454–1459. doi: 10.1038/bjc.1998.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee SH, Shin MS, Park WS, Kim SY, Dong SM, Pi JH, Lee HK, Kim HS, Jang JJ, Kim CS, et al. Alterations of Fas (APO-1/CD95) gene in transitional cell carcinomas of urinary bladder. Cancer Res. 1999;59:3068–3072. [PubMed] [Google Scholar]
- 29.Shimonishi T, Isse K, Shibata F, Aburatani I, Tsuneyama K, Sabit H, Harada K, Miyazaki K, Nakanuma Y. Up-regulation of fas ligand at early stages and down-regulation of Fas at progressed stages of intrahepatic cholangiocarcinoma reflect evasion from immune surveillance. Hepatology. 2000;32:761–769. doi: 10.1053/jhep.2000.18192. [DOI] [PubMed] [Google Scholar]
- 30.Huang Q, Wang J, Hu Y. FAS-670 gene polymorphism and cervical carcinogenesis risk: A meta-analysis. Biomed Rep. 2013;1:889–894. doi: 10.3892/br.2013.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38:209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 32.Solé X, Guinó E, Valls J, Iniesta R, Moreno V. SNPStats: A web tool for the analysis of association studies. Bioinformatics. 2006;22:1928–1929. doi: 10.1093/bioinformatics/btl268. [DOI] [PubMed] [Google Scholar]
- 33.Ziegler A, Van Steen K, Wellek S. Investigating Hardy-Weinberg equilibrium in case-control or cohort studies or meta-analysis. Breast Cancer Res Treat. 2011;128:197–201. doi: 10.1007/s10549-010-1295-z. [DOI] [PubMed] [Google Scholar]
- 34.Narita K, Staub J, Chien J, Meyer K, Bauer M, Friedl A, Ramakrishnan S, Shridhar V. HSulf-1 inhibits angiogenesis and tumorigenesis in vivo. Cancer Res. 2006;66:6025–6032. doi: 10.1158/0008-5472.CAN-05-3582. [DOI] [PubMed] [Google Scholar]
- 35.Lai J, Chien J, Staub J, Avula R, Greene EL, Matthews TA, Smith DI, Kaufmann SH, Roberts LR, Shridhar V. Loss of HSulf-1 up-regulates heparin-binding growth factor signaling in cancer. J Biol Chem. 2003;278:23107–23117. doi: 10.1074/jbc.M302203200. [DOI] [PubMed] [Google Scholar]
- 36.Zhou Q, Jiang Y, Yin W, Wang Y, Lu J. Single-nucleotide polymorphism in microRNA-binding site of SULF1 target gene as a protective factor against the susceptibility to breast cancer: A case-control study. Onco Targets Ther. 2016;9:2749–2757. doi: 10.2147/OTT.S102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okolicsanyi RK, Faure M, Jacinto JM, Chacon-Cortes D, Chambers S, Youl PH, Haupt LM, Griffiths LR. Association of the SNP rs2623047 in the HSPG modification enzyme SULF1 with an Australian Caucasian breast cancer cohort. Gene. 2014;547:50–54. doi: 10.1016/j.gene.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 38.Han CH, Huang YJ, Lu KH, Liu Z, Mills GB, Wei Q, Wang LE. Polymorphisms in the SULF1 gene are associated with early age of onset and survival of ovarian cancer. J Exp Clin Cancer Res. 2011;30:5. doi: 10.1186/1756-9966-30-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tamandani Kordi DM, Sobti RC, Shekari M. Association of Fas-670 gene polymorphism with risk of cervical cancer in North Indian population. Clin Exp Obstet Gynecol. 2008;35:183–186. [PubMed] [Google Scholar]
- 40.Ueda M, Terai Y, Kanda K, Kanemura M, Takehara M, Yamaguchi H, Nishiyama K, Yasuda M, Ueki M. Fas gene promoter −670 polymorphism in gynecological cancer. Int J Gynecol Cancer. 2006;16(Suppl 1):179–182. doi: 10.1111/j.1525-1438.2006.00505.x. [DOI] [PubMed] [Google Scholar]
- 41.Nunobiki O, Ueda M, Toji E, Yamamoto M, Akashi K, Sato N, Izuma S, Torii K, Tanaka I, Okamoto Y, et al. Genetic polymorphism of cancer susceptibility genes and HPV infection in cervical carcinogenesis. Pathol Res Int. 2011;2011:364069. doi: 10.4061/2011/364069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lai HC, Lin WY, Lin YW, Chang CC, Yu MH, Chen CC, Chu TY. Genetic polymorphisms of FAS and FASL (CD95/CD95L) genes in cervical carcinogenesis: An analysis of haplotype and gene-gene interaction. Gynecol Oncol. 2005;99:113–118. doi: 10.1016/j.ygyno.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 43.Lai HC, Sytwu HK, Sun CA, Yu MH, Yu CP, Liu HS, Chang CC, Chu TY. Single nucleotide polymorphism at Fas promoter is associated with cervical carcinogenesis. Int J Cancer. 2003;103:221–225. doi: 10.1002/ijc.10800. [DOI] [PubMed] [Google Scholar]
- 44.Tan SC, Ismail MP, Duski DR, Othman NH, Ankathil R. FAS c.-671A>G polymorphism and cervical cancer risk: A case-control study and meta-analysis. Cancer Genet. 2017;211:18–25. doi: 10.1016/j.cancergen.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 45.Hammond E, Khurana A, Shridhar V, Dredge K. The role of heparanase and sulfatases in the modification of heparan sulfate proteoglycans within the tumor microenvironment and opportunities for novel cancer therapeutics. Front Oncol. 2014;4:195. doi: 10.3389/fonc.2014.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hur K, Han TS, Jung EJ, Yu J, Lee HJ, Kim WH, Goel A, Yang HK. Up-regulated expression of sulfatases (SULF1 and SULF2) as prognostic and metastasis predictive markers in human gastric cancer. J Pathol. 2012;228:88–98. doi: 10.1002/path.4055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.