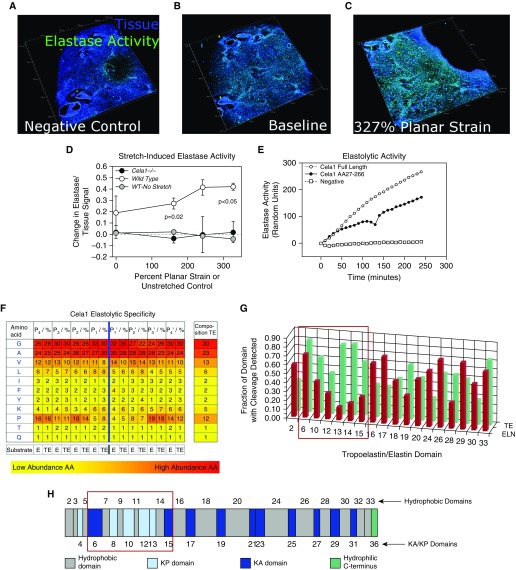
Figure 2.
Elastolytic profile of Cela1. (A) Live lung sections of 12-week-old mice were inflated with gelatin, then 200-μm sections were adhered to the silicone insert of a three-dimensional printed confocal lung-stretching device (see Figure E3), and the gelatin was removed. Lung tissue was defined by autofluorescence, and elastase activity was determined using a soluble elastin substrate conjugated to a quenched fluorophore. A three-dimensional image of a lung section is shown, with each major tick mark representing 500 μm. (B) After application of the zymography substrate, there was an appreciable increase in the elastase signal. A representative WT lung section is shown. (C) The same lung section is shown at the maximal strain with an increase in the elastase signal. (D) The stretch-induced lung elastase activity of WT (n = 3) and Cela1−/− (n = 2) lung sections was compared. WT lung not exposed to stretch but incubated with substrate for an equivalent time did not demonstrate increased elastase signal. Cela1−/− lung that was stretched lacked the inducible elastase activity observed in WT lung. Comparisons were carried out using one-way ANOVA. (E) As assessed using a plate-based fluorometric elastase assay, full-length Cela1 demonstrated elastolytic activity comparable to that of Cela1 without signaling and propeptide. (F) Full-length recombinant Cela1 was incubated with soluble human tropoelastin and human skin elastin, and degradation products were analyzed to determine proteolytic specificity. Whereas Cela1 had a propensity for hydrophobic residues, the only amino acid that was nonpreferred in regions adjacent to cleavage sites was proline. However, proline was preferred at P4-P2 and P3′-P4′ sites, suggesting that the conformational turn induced by proline residues at these locations enhanced elastin–Cela1 interaction. (G) Although there was no major difference in the amino acid residues cleaved by Cela1, mature elastin had substantially fewer cleavage sites in domains 6–15 (red box). (H) Although there was no major difference in the hydrophobicity of these domains (ivory color, domains numbered above simplified tropoelastin protein sequence) compared with others, these domains contain lysine-proline (KP) cross-linking sites (light blue color). Because soluble tropoelastin lacks these cross-links, they likely account for the difference in elastin degradation between mature elastin and soluble tropoelastin. AA = amino acid; E, ELN = elastin; KA = lysine-alanine; TE = tropoelastin.